Roles for mycobacterial DinB2 in frameshift and substitution mutagenesis
Figures
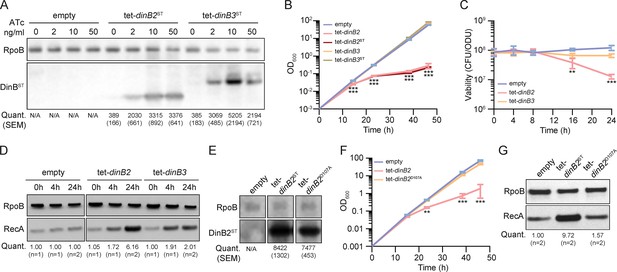
Overexpression of DinB2 causes cell death through its polymerase activity.
(A) Anti-streptavidin/RpoB immunoblots from indicated strains with indicated concentrations of inducer treatment (16 hr of treatment). Average and SEM of RpoB normalized band intensities (n=3, arbitrary units) are given below the image of a representative blot. (B) Growth and (C) viability of indicated strains in presence of 50 nM anhydrotetracycline (ATc). Note that the OD600 values in (B) are calculated values based on continuous dilution growth experiments (see Methods). (D) Anti-RecA/RpoB immunoblots from indicated strains with indicated times of inducer treatment (50 nM). Average of normalized band intensities, expressed relative to the empty vector strain, is given below the image of a representative blot. (E) Anti-streptavidin/RpoB immunoblots from indicated strains after 16 hr of inducer treatment (50 nM ATc). Average and SEM of normalized band intensities (n=3) are given below the image of a representative blot. (F) Growth of indicated strains in presence of 50 nM ATc. (G) Anti-RecA/RpoB immunoblots from indicated strains after 24 hr of inducer treatment. Average of normalized band intensities, expressed relatively to the empty vector strain, is given below the image of a representative blot. Empty = empty vector, tet = ATc-inducible promoter, DinB2=M. smegmatis DinB2, DinB3=M. smegmatis DinB3, ST=streptavidin tag, D107A=catalytically inactive M. smegmatis DinB2. Results shown are means (± SEM) of biological triplicates. Stars under the means mark a statistical difference compared to the empty vector reference strain (**, p<0.01; ***, p<0.001).
-
Figure 1—source data 1
Uncropped immunoblots Figure 1A, D, E and G.
- https://cdn.elifesciences.org/articles/83094/elife-83094-fig1-data1-v1.zip

DinB2 overexpression induces growth defect in M. smegmatis.
(A) Anti-Streptavidin/RpoB immunoblots from indicated strains with indicated times of inducer treatment (50 nM ATc). (B and D) Growth of indicated strains of M. smegmatis on agar medium containing the indicated concentrations of inducer (ATc) in agar. (C) Liquid growth of M. smegmatis carrying the dinB2 expression plasmid in presence of the indicated concentrations of ATc. Results shown are means (± SEM) of data obtained from biological triplicates. Stars above or under the means mark a statistical difference with the reference strain (0nM of inducer) (**, P<0.01; ***, P<0.001). Empty=empty vector, tet=Atc inducible promoter, DinB2=M. smegmatis DinB2, DinB3=M. smegmatis DinB3, ST=Streptavidin tag, D107A=catalytically inactive M. smegmatis DinB2, L14F=Steric gate mutant of M. smegmatis DinB2.
-
Figure 1—figure supplement 1—source data 1
Uncropped immunoblots Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/83094/elife-83094-fig1-figsupp1-data1-v1.zip
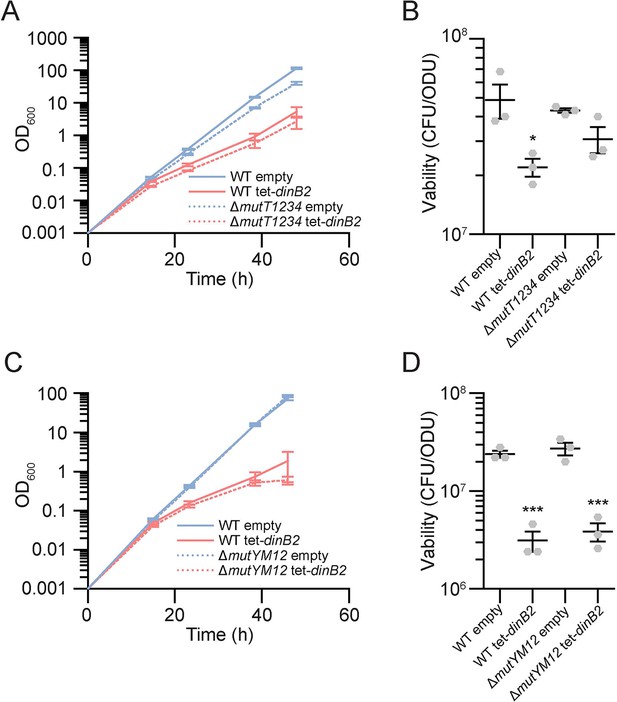
Lethality of DinB2 overexpression in absence of anti-8-oxoguanine systems.
(A and C) Growth and (B and D) viability of indicated strains in presence of 50 nM ATc. Empty=empty vector, tet=Atc inducible promoter, DinB2=M. smegmatis DinB2, DinB3=M. smegmatis DinB3. Results shown are means (± SEM) of data obtained from biological triplicates. Stars above means mark a statistical difference with the empty vector (*, P<0.05; ***, P<0.001).
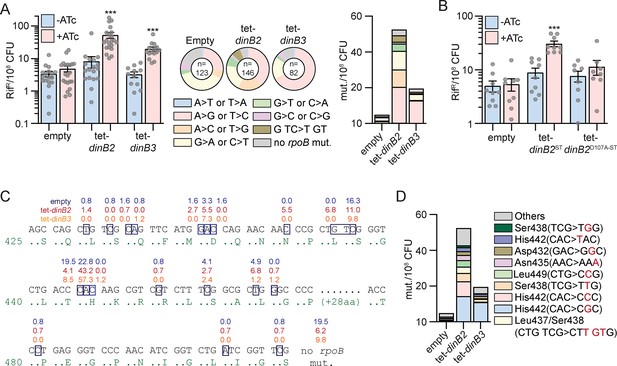
DinB2 and DinB3 overexpression confers antibiotic resistance through a distinct mutagenic profile.
(A and B) Rifampicin resistance (rifR) frequency in indicated strains in absence (blue) or presence (red) of inducer (50 nM anhydrotetracycline [ATc]). Results shown are means (± SEM) of data obtained from biological replicates symbolized by gray dots. Stars above bars mark a statistical difference with the reference (same strain without inducer) (***, p<0.001). Pie charts and bar chart in (A) shows the relative and absolute frequencies of nucleotide changes, represented with colors, detected in rpoB of indicated strains rifR in presence of inducer (50 nM ATc). The number of sequenced rifR is given in the center of each pie chart. (C) Location and relative frequency in % of mutated nucleotides in rpoB found in empty (blue), tet-dinB2 (red), or tet-dinB3 (orange) rifR. (D) Absolute frequency of the main rpoB mutations found in indicated strains in presence of 50 nM ATc. Empty = empty vector, tet = ATc-inducible promoter, DinB2=M. smegmatis DinB2, DinB3=M. smegmatis DinB3, ST = streptavidin tag, D107A=catalytically inactive M. smegmatis DinB2.

Mutation frequency in ΔrecA and ΔdnaE2 backgrounds after DinB2 and DinB3 overexpression.
Rifampicin resistance (rifR) frequency after DinB2 or DinB3 overexpression in (A) ΔrecA or (B) ΔdnaE2 backgrounds in absence (blue) or presence (red) of inducer (ATc 50 nM; 16h of treatment). Results shown are means (± SEM) of data obtained from biological replicates symbolized by grey dots. Stars above or under means mark a statistical difference with the same strain without inducer (*, P<0.05; ***, P<0.001).
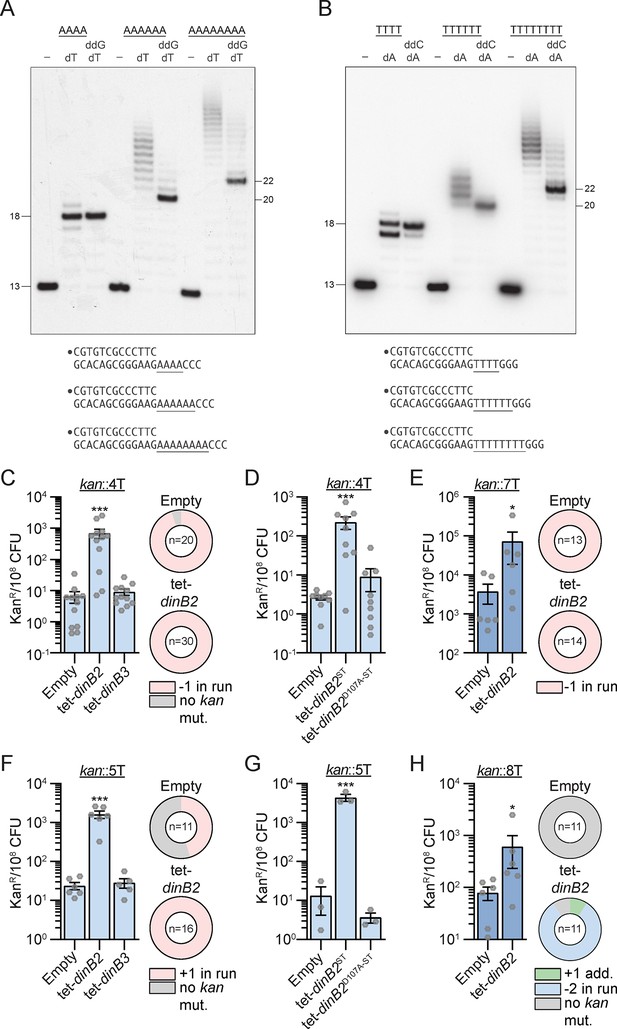
DinB2 efficiently promotes –1 and +1 frameshifts in short and long runs of A and T.
(A and B) Reaction mixtures containing 10 mM Tris-HCl, pH 7.5, 5 mM MnCl2,1 pmol 5' 32P-labeled primer-template DNAs with indicated runs in the template strand (depicted below, and included as indicated above the lanes), 125 µM dTTP and ddGTP as specified, and 10 pmol DinB2 were incubated at 37°C for 15 min. The reaction products were analyzed by urea-PAGE and visualized by autoradiography. DinB2 was omitted from reactions in lanes –. (C–H) kanR frequencies in the indicated strains carrying the indicated mutation reporters in presence of inducer (50 nM ATc). Results shown are means (± SEM) of data obtained from biological replicates symbolized by gray dots. Stars above the bars mark a statistical difference with the reference strain (empty) (*, p<0.05; ***, p<0.001). Relative (pie chart) and absolute (bar chart) frequencies of nucleotide changes detected in kan of kanR cells represented with colors: pink=–1 or +1 frameshift in the homo-oligonucleotide run, blue=-2 frameshift in the run, green=+1 frameshift localized outside of the run and gray=no detected mutation. The number of sequenced kanR colonies is given in the center of each pie chart. Empty=empty vector, tet = Atc-inducible promoter, DinB2=M. smegmatis DinB2, DinB3=M. smegmatis DinB3, ST=streptavidin tag, D107A=catalytically inactive M. smegmatis DinB2.
-
Figure 3—source data 1
Original autoradiograms (Figure 3A and B).
- https://cdn.elifesciences.org/articles/83094/elife-83094-fig3-data1-v1.zip
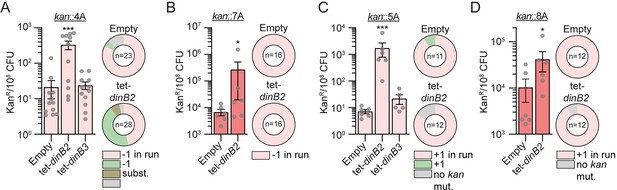
Frameshift mutagenesis in diverse runs after DinB2 and DinB3 overexpression.
KanR frequencies in the indicated strains carrying indicated mutation reporters (kan::4A (A), kan::7A (B), kan::5A (C), kan::8A (D), kan::7G (E), and kan::8G (F)) in presence of ATc 50 nM. Results shown are means (± SEM) of data obtained from biological replicates symbolized by grey dots. Stars above the means mark a statistical difference with the reference strain (empty) (*, P<0.05; ***, P<0.001). Relative (pie chart) and absolute (bar chart) frequencies of nucleotide changes detected in kan of kanR cells represented with colors: pink=-1 or +1 frameshift in the homo-oligonucleotide run, green=-1 or +1 frameshift localized outside of the run, brown=substitution mutations, and grey=no detected mutation. The number of sequenced kanR colonies is given in the center of each pie chart. Empty=empty vector, tet=Atc inducible promoter, DinB2=M. smegmatis DinB2, DinB3=M. smegmatis DinB3.
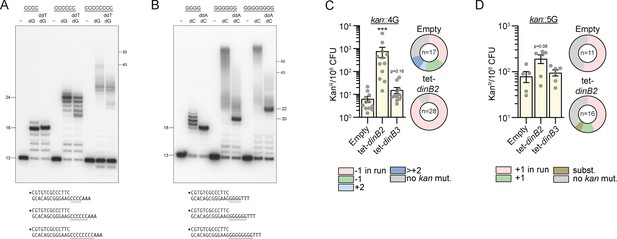
DinB2 slippage activity is enhanced on C and G homo-oligonucleotide templates.
(A and B) Reaction mixtures containing 10 mM Tris-HCl, pH 7.5, 5 mM MnCl2,1 pmol 5' 32P-labeled primer-template DNAs with indicated runs in the template strand (depicted below, and included as indicated above the lanes), 125 µM dGTP and ddTTP or dCTP and ddATP as specified, and 10 pmol DinB2 were incubated at 37°C for 15 min. DinB2 was omitted from reactions in lanes –. The reaction products were analyzed by urea-PAGE and visualized by autoradiography. The positions of the 13-mer primer strand and 5' 32P-labeled 40-mer and 50-mer oligonucleotide size markers analyzed in parallel are indicated on the right. (C–D) kanR frequencies in the indicated strains carrying indicated mutation reporters in presence of inducer (50 nM anhydrotetracyclin [ATc]). Results shown are means (± SEM) of data obtained from biological replicates symbolized by gray dots. Stars above the means mark a statistical difference with the reference strain (empty) (***, p<0.001). Relative (pie chart) and absolute (bar chart) frequencies of nucleotide changes detected in kan of kanR cells represented with colors: pink=–1 or +1 frameshift (FS) in the homo-oligonucleotide run, green=–1 or +1 FS localized outside of the run, light blue=+2 FS localized outside of the run, dark blue=>+2 insertion, brown=bases substitution mutation and gray=no detected mutation. The number of sequenced kanR colonies is given in the center of each pie chart. Empty=empty vector, tet=Atc-inducible promoter, DinB2=M. smegmatis DinB2, DinB3=M. smegmatis DinB3.
-
Figure 4—source data 1
Original autoradiograms (Figure 4A and B).
- https://cdn.elifesciences.org/articles/83094/elife-83094-fig4-data1-v1.zip
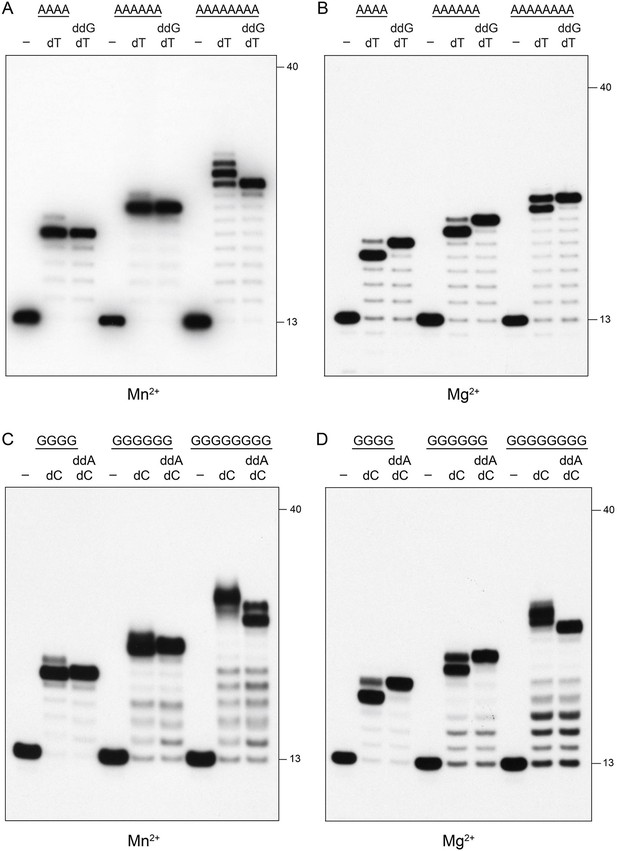
Pol1 is not prone to slippage.
Reaction mixtures containing 10 mM Tris-HCl, pH 7.5, 5 mM MnCl2 or MgCl2 as specified below each panel, 1 pmol 5' 32P-labeled primer-template DNAs with A4, A6, A8, G4, G6, or G8 runs in the template strand (included as indicated above the lanes), nucleotides as specified, and 10 pmol Pol1 POL domain were incubated at 37°C for 15 min. Pol1 was omitted from reactions in lanes –. The reaction products were analyzed by urea-PAGE and visualized by autoradiography. The positions of the 13-mer primer strand and a 5' 32P-labeled 40-mer oligonucleotide size markers analyzed in parallel are indicated on the right.
-
Figure 4—figure supplement 1—source data 1
Original autoradiograms (Figure 4—figure supplement 1A–D).
- https://cdn.elifesciences.org/articles/83094/elife-83094-fig4-figsupp1-data1-v1.zip

DinB2 does not incorporate long slippage products in vivo.
SucR frequencies in the indicated strains carrying indicated mutation reporters in presence or absence of inducer Results shown are means (± SEM) of data obtained from biological replicates symbolized by grey dots. Stars above the means mark a statistical difference with the reference strain (empty) (**, P<0.01; ***, P<0.001). Relative frequencies of nucleotide changes detected in sacB of sucR cells are represented with colors: dark red = +1 frameshift in the homo-oligonucleotide run, light red = -1 frameshift in the homo-oligonucleotide run, green=-1 frameshift localized outside of the run, brown=substitution mutations, and grey=no detected mutation. The number of sequenced sucR colonies is given in the center of each pie chart. Empty=empty vector, tet=Atc inducible promoter, DinB2=M. smegmatis DinB2.
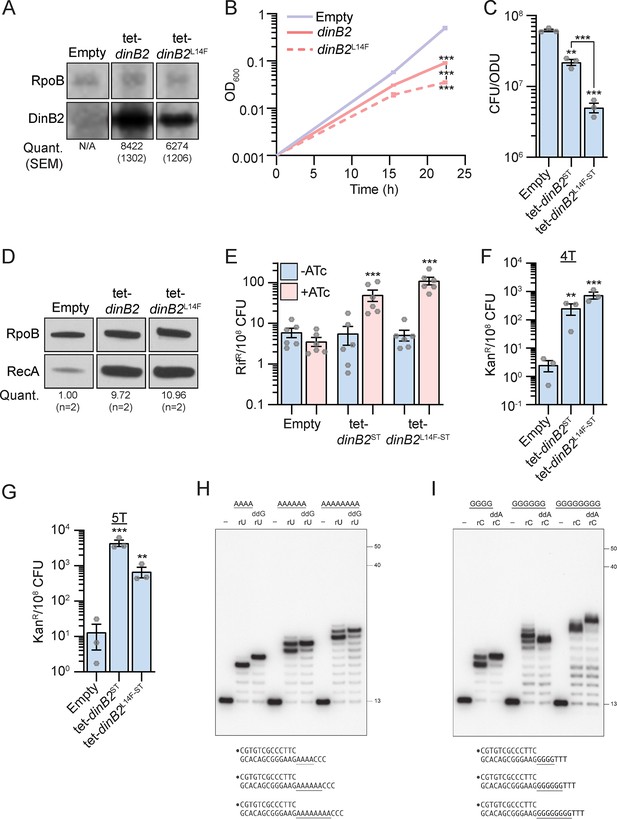
DinB2 does not slip in RNA polymerase mode.
(A) Anti-streptavidin/RpoB immunoblots from indicated strains after 16 hr of inducer treatment (50 nM anhydrotetracycline [ATc]). Average and SEM of RpoB normalized band intensities (n=3) are given below the image of a representative blot. (B) Growth of indicated strains in presence of 50 nM ATc. (C) Viability of indicated strains after 24 hr of inducer treatment (50 nM ATc). (D) Anti-RecA/RpoB immunoblots from indicated strains after 24 hr of inducer treatment (50 nM ATc). Average of normalized band intensities, expressed relative to the empty condition, is given below the image of a representative blot. (E) Rifampicin resistance (rifR) frequency in indicated strains in absence (blue) or presence (pink) of 50 nM ATc. (F and G) kanR frequencies in the indicated strains carrying indicated mutation reporters in presence of 50 nM ATc. Results shown are means (± SEM) of data obtained from biological replicates symbolized by gray dots or biological triplicates for (B). Stars above the means mark a statistical difference with the reference strain (B, C, F, and G: empty or E: same strain without inducer). Lines connecting two strains show a statistical difference between them. (**, p<0.01; ***, p<0.001). (H and I) Reaction mixtures containing 10 mM Tris-HCl, pH 7.5, 5 mM MnCl2, 1 pmol 5' 32P-labeled primer-template DNAs with indicated runs in the template strand (depicted below, and included as indicated above the lanes), 125 µM rUTP and ddGTP or rCTP and ddATP as specified, and 10 pmol DinB2 were incubated at 37°C for 15 min. DinB2 was omitted from reactions in lanes –. The reaction products were analyzed by urea-PAGE and visualized by autoradiography. The positions of the 13-mer primer strand and 5' 32P-labeled 40-mer and 50-mer oligonucleotide size markers analyzed in parallel are indicated on the right. Empty=empty vector, tet=Atc-inducible promoter, DinB2=M. smegmatis DinB2, ST=streptavidin tag, L14F=steric gate mutant of M. smegmatis DinB2.
-
Figure 5—source data 1
Uncropped immunoblots (Figure 5A and D) and original autoradiograms (Figure 5H and I).
- https://cdn.elifesciences.org/articles/83094/elife-83094-fig5-data1-v1.zip
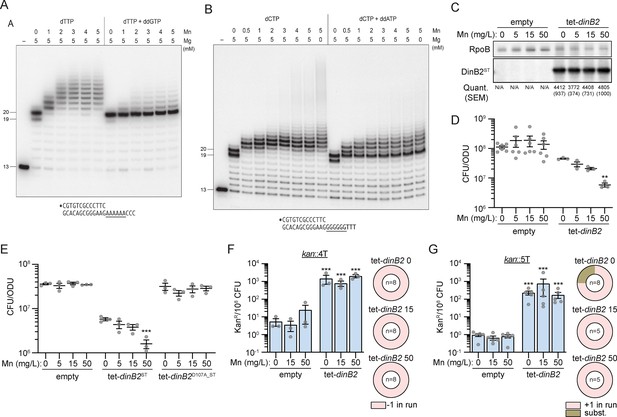
Metal-dependent switch in DinB2 activities.
(A and B) Reaction mixtures containing 10 mM Tris-HCl, pH 7.5, 1 pmol 5' 32P-labeled primer-template DNAs with an A6 or G6 run in the template strand (depicted below), divalent cations and nucleotides (125 µM) as specified above the lanes, and 10 pmol DinB2 were incubated at 37°C for 15 min. DinB2 was omitted from reactions in lanes –. The reaction products were analyzed by urea-PAGE and visualized by autoradiography. (C) Anti-streptavidin/RpoB immunoblots from indicated strains cultivated with indicated concentrations of MnCl2 (in mg/L) after 16 hr of inducer treatment (50 nM anhydrotetracyclin [ATc]). Average and SEM of normalized band intensities (n=3) are given below the image of representative blot. (D and E) Viability of indicated strains after 24 hr of inducer treatment (50 nM ATc) in presence of indicated concentration of MnCl2 (in mg/L). (F and G) kanR frequencies in the indicated strains carrying indicated mutation reporters in presence of inducer (50 nM ATc) and indicated concentration of MnCl2 (in mg/L). Results shown are means (± SEM) of data obtained from biological replicates symbolized by gray dots. Relative frequencies of nucleotide changes detected in kan of kanR cells are represented with colors: pink=–1 or +1 frameshift in the homo-oligonucleotide run, brown = substitution mutations. The number of sequenced kanR colonies is given in the center of each pie chart. Stars above the means mark a statistical difference with the reference strain: same strain untreated with Mn in (D) and (E), empty with same Mn treatment in (F) and (G) (**, p<0.01; ***, p<0.001). Empty = empty vector, tet = Atc-inducible promoter, DinB2=M. smegmatis DinB2, ST = streptavidin tag, D107A=catalytically inactive M. smegmatis DinB2.
-
Figure 6—source data 1
Original autoradiograms (Figure 6A and B) and uncropped immunoblots (Figure 6C).
- https://cdn.elifesciences.org/articles/83094/elife-83094-fig6-data1-v1.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | DH5α | The Glickman lab | ||
| Strain, strain background (Mycobacterium smegmatis) | Wild type (mc2155) | Snapper et al., 1990 | PDS1 | |
| Strain, strain background (Mycobacterium smegmatis) | ∆recA | Dupuy et al., 2020 | PDS353 | |
| Strain, strain background (Mycobacterium smegmatis) | ∆dnaE2 | Dupuy et al., 2020 | PDS139 | |
| Strain, strain background (Mycobacterium smegmatis) | pmsg419 | This work | Mgm4062 | Available from Glickman Lab |
| Strain, strain background (Mycobacterium smegmatis) | pRGM47 | This work | mgm4063 | Available from Glickman Lab |
| Strain, strain background (Mycobacterium smegmatis) | pRGM48 | This work | mgm4072 | Available from Glickman Lab |
| Strain, strain background (Mycobacterium smegmatis) | pRGM49 | This work | mgm4073 | Available from Glickman Lab |
| Strain, strain background (Mycobacterium smegmatis) | pRGM50 | This work | mgm4074 | Available from Glickman Lab |
| Strain, strain background (Mycobacterium smegmatis) | pDP69 | This work | PDS416 | Available from Glickman Lab |
| Strain, strain background (Mycobacterium smegmatis) | pDP70 | This work | PDS417 | Available from Glickman Lab |
| Peptide, recombinant protein | DinB1 | Ordonez et al., 2014 | ||
| Peptide, recombinant protein | DinB2 | Ordonez et al., 2014 | ||
| Recombinant DNA reagent | ATc-on system vector (hygR, OriMyc) | Lab Stock | pmsg419 | Available from Glickman Lab |
| Recombinant DNA reagent | Mycob. integr. vector (StrepR, attP(L5)) | Lab Stock | pDB60 | Available from Glickman Lab |
| Recombinant DNA reagent | pmsg419-dinB2 Strep tag | This work | pRGM47 | Cloning primers: dinB2fw-dinB2rev1 (cloning enzyme site: ClaI); available from Glickman Lab |
| Recombinant DNA reagent | pmsg419-dinB2 | This work | pRGM48 | Cloning primers: dinB2fw-dinB2rev2 (cloning enzyme site: ClaI); dinB2=Msmeg_2294; available from Glickman Lab |
| Recombinant DNA reagent | pmsg419-dinB2D107A Strep tag | This work | pRGM49 | Cloning primers: dinB2fw-dinB2catrev +dinB2catfw-dinB2rev1 (cloning enzyme site: ClaI); available from Glickman Lab |
| Recombinant DNA reagent | pmsg419-dinB2L14F Strep tag | This work | pRGM50 | Cloning primers: dinB2fw-dinB2stericrev +dinB2stericfw-dinB2rev1 (cloning enzyme site: ClaI); available from Glickman Lab |
| Recombinant DNA reagent | pmsg419-dinB3 | This work | pDP69 | Cloning primers: ODP197-ODP298 (cloning enzyme site: ClaI); available from Glickman Lab |
| Recombinant DNA reagent | pmsg419-dinB3 Strep tag | This work | pDP70 | Cloning primers: ODP197-ODP299 (cloning enzyme site: ClaI); available from Glickman Lab |
| Recombinant DNA reagent | pDB60 derivative with kan::3T | Dupuy et al., 2022 | pDP120 | |
| Recombinant DNA reagent | pDB60 derivative with kan::3C | Dupuy et al., 2022 | pDP121 | |
| Recombinant DNA reagent | pDB60 derivative with kan::3G | Dupuy et al., 2022 | pDP122 | |
| Recombinant DNA reagent | pDB60 derivative with kan::3A | Dupuy et al., 2022 | pDP123 | |
| Recombinant DNA reagent | pDB60 derivative with kan::4T | Dupuy et al., 2022 | pDP124 | |
| Recombinant DNA reagent | pDB60 derivative with kan::4C | Dupuy et al., 2022 | pDP125 | |
| Recombinant DNA reagent | pDB60 derivative with kan::4G | Dupuy et al., 2022 | pDP126 | |
| Recombinant DNA reagent | pDB60 derivative with kan::4A | Dupuy et al., 2022 | pDP127 | |
| Recombinant DNA reagent | pDB60 derivative with kan::6T | Dupuy et al., 2022 | pDP128 | |
| Recombinant DNA reagent | pDB60 derivative with kan::6C | Dupuy et al., 2022 | pDP129 | |
| Recombinant DNA reagent | pDB60 derivative with kan::6G | Dupuy et al., 2022 | pDP130 | |
| Recombinant DNA reagent | pDB60 derivative with kan::6A | Dupuy et al., 2022 | pDP131 | |
| Recombinant DNA reagent | pDB60 derivative with kan::7T | This work | pDP132 | Cloning primers: ODP443-ODP445+ODP458-ODP444 (cloning enzyme site: EcoRI); available from Glickman Lab |
| Recombinant DNA reagent | pDB60 derivative with kan::7C | This work | pDP133 | Cloning primers: ODP443-ODP445+ODP459-ODP444 (cloning enzyme site: EcoRI); available from Glickman Lab |
| Recombinant DNA reagent | pDB60 derivative with kan::7G | This work | pDP134 | Cloning primers: ODP443-ODP445+ODP460-ODP444 (cloning enzyme site: EcoRI); available from Glickman Lab |
| Recombinant DNA reagent | pDB60 derivative with kan::7A | This work | pDP135 | Cloning primers: ODP443-ODP445+ODP461-ODP444 (cloning enzyme site: EcoRI); available from Glickman Lab |
| Recombinant DNA reagent | pDB60 derivative with kan::9T | This work | pDP136 | Cloning primers: ODP443-ODP445+ODP462-ODP444 (cloning enzyme site: EcoRI); available from Glickman Lab |
| Recombinant DNA reagent | pDB60 derivative with kan::9C | This work | pDP137 | Cloning primers: ODP443-ODP445+ODP463-ODP444 (cloning enzyme site: EcoRI); available from Glickman Lab |
| Recombinant DNA reagent | pDB60 derivative with kan::9G | This work | pDP138 | Cloning primers: ODP443-ODP445+ODP464-ODP444 (cloning enzyme site: EcoRI); available from Glickman Lab |
| Recombinant DNA reagent | pDB60 derivative with kan::9A | This work | pDP139 | Cloning primers: ODP443-ODP445+ODP465-ODP444 (cloning enzyme site: EcoRI); available from Glickman Lab |
| Recombinant DNA reagent | pDB60 derivative with kan::5T | Dupuy et al., 2022 | pDP144 | |
| Recombinant DNA reagent | pDB60 derivative with kan::5C | Dupuy et al., 2022 | pDP145 | |
| Recombinant DNA reagent | pDB60 derivative with kan::5G | Dupuy et al., 2022 | pDP146 | |
| Recombinant DNA reagent | pDB60 derivative with kan::5A | Dupuy et al., 2022 | pDP147 | |
| Recombinant DNA reagent | pDB60 derivative with kan::8T | This work | pDP148 | Cloning primers: ODP443-ODP445+ODP494-ODP444 (cloning enzyme site: EcoRI); available from Glickman Lab |
| Recombinant DNA reagent | pDB60 derivative with kan::8C | This work | pDP149 | Cloning primers: ODP443-ODP445+ODP495-ODP444 (cloning enzyme site: EcoRI); available from Glickman Lab |
| Recombinant DNA reagent | pDB60 derivative with kan::8G | This work | pDP150 | Cloning primers: ODP443-ODP445+ODP496-ODP444 (cloning enzyme site: EcoRI); available from Glickman Lab |
| Recombinant DNA reagent | pDB60 derivative with kan::8A | This work | pDP151 | Cloning primers: ODP443-ODP445+ODP497-ODP444 (cloning enzyme site: EcoRI); available from Glickman Lab |
| Recombinant DNA reagent | pDB60 derivative with sacB::9C | This work | pDP186 | Cloning primers: ODP593-ODP596+ODP597-ODP598 (cloning enzyme site: EcoRI); available from Glickman Lab |
| Recombinant DNA reagent | pDB60 derivative with sacB::6C | This work | pDP194 | Cloning primers: ODP593-ODP614+ODP615-ODP598 (cloning enzyme site: EcoRI); available from Glickman Lab |
| Antibody | Anti-RpoB (mouse monoclonal) | Biolegend | 663905; AB_2566583 | (1:10,000 dilution) |
| Antibody | Anti-RecA (rabbit polyclonal) | Wipperman et al., 2018 | Anti-RecA | (1:10,000 dilution) |
| Antibody | Anti-streptavidin (rabbit polyclonal) | GenScript | A00626 | STII GenScript rabbit anti-NWSHPQFEK; used at (1:10,000 dilution) |
| Sequence-based reagent | fw dinB2 | IDT | dinB2fw | CAGAAAGGAGGCCATATGACCAAATGGGTGCTC |
| Sequence-based reagent | rev dinB2+streptavidin tag | IDT | dinB2rev1 | AGGTCGACGGTATCGATACTACTTTTCGAACTGCG GGTGGCTCCAGGTGCCTGCAGTGACAG |
| Sequence-based reagent | rev dinB2 | IDT | dinB2rev2 | AGGTCGACGGTATCGATGTGCTCGAGTTAGGTGCCTGCAGTGAC |
| Sequence-based reagent | rev internal dinB2Msm with pol. dead mut. (D107A) | IDT | dinB2catrev | GCCCAGATACGCCTCGGCCCAGCCCCACACCTCCAAC |
| Sequence-based reagent | fw internal dinB2Msm with pol. dead mut. (D107A) | IDT | dinB2catfw | GCCGAGGCGTATCTGGGC |
| Sequence-based reagent | rev internal dinB2Msm with steric gate mut. (L14F) | IDT | dinB2stericrev | GCAACTCCACCGAAGCAAAGAACTGGTCCAGATCGAC |
| Sequence-based reagent | fw internal dinB2Msm with steric gate mut. (L14F) | IDT | dinB2stericfw | TTTGCTTCGGTGGAGTTGC |
| Sequence-based reagent | fw dinB3 | IDT | ODP297 | CAGAAAGGAGGCCATATGTTCGTGTCCGCTGC |
| Sequence-based reagent | rev dinB3 | IDT | ODP298 | AGGTCGACGGTATCGCTAGTCCGGCAGCATGG |
| Sequence-based reagent | rev dinB3+streptavidin tag | IDT | ODP299 | AGGTCGACGGTATCGCTACTTTTCGAACTGCGGGT GGCTCCAGTCCGGCAGCATGGG |
| Sequence-based reagent | fw kan | IDT | ODP443 | TCCAGCTGCAGAATTTCCCAAGGACACTGAGTCC |
| Sequence-based reagent | rev kan | IDT | ODP444 | GATAAGCTTCGAATTTTGCTGACTCATACCAGGC |
| Sequence-based reagent | Internal rev kan | IDT | ODP445 | CATAACACCCCTTGTATTACTG |
| Sequence-based reagent | Internal fw kan (7T addition) | IDT | ODP458 | ACAAGGGGTGTTATGTTTTTTTAGCCATATTCAACGGGAAACG |
| Sequence-based reagent | Internal fw kan (7C addition) | IDT | ODP459 | ACAAGGGGTGTTATGCCCCCCCAGCCATATTCAACGGGAAACG |
| Sequence-based reagent | Internal fw kan (7G addition) | IDT | ODP460 | ACAAGGGGTGTTATGGGGGGGAAGCCATATTCAACGGGAAACG |
| Sequence-based reagent | Internal fw kan (7A addition) | IDT | ODP461 | ACAAGGGGTGTTATGGAAAAAAAGCCATATTCAACGGGAAACG |
| Sequence-based reagent | Internal fw kan (9T addition) | IDT | ODP462 | ACAAGGGGTGTTATGTTTTTTTTTAGCCATATTCAACGGGAAACG |
| Sequence-based reagent | Internal fw kan (9C addition) | IDT | ODP463 | ACAAGGGGTGTTATGCCCCCCCCCAGCCATATTCAACGGGAAACG |
| Sequence-based reagent | Internal fw kan (9G addition) | IDT | ODP464 | ACAAGGGGTGTTATGGGGGGGGGAAGCCATATTCAACGGGAAACG |
| Sequence-based reagent | Internal fw kan (9A addition) | IDT | ODP465 | ACAAGGGGTGTTATGGAAAAAAAAAGCCATATTCAACGGGAAACG |
| Sequence-based reagent | Internal fw kan (8T addition) | IDT | ODP494 | ACAAGGGGTGTTATGTTTTTTTTAGCCATATTCAACGGGAAACG |
| Sequence-based reagent | Internal fw kan (8C addition) | IDT | ODP495 | ACAAGGGGTGTTATGCCCCCCCCAGCCATATTCAACGGGAAACG |
| Sequence-based reagent | Internal fw kan (8G addition) | IDT | ODP496 | ACAAGGGGTGTTATGGGGGGGGAAGCCATATTCAACGGGAAACG |
| Sequence-based reagent | Internal fw kan (8A addition) | IDT | ODP497 | ACAAGGGGTGTTATGGAAAAAAAAGCCATATTCAACGGGAAACG |
| Sequence-based reagent | fw sacB | IDT | ODP593 | TCCAGCTGCAGAATTAACCCATCACATATACCTGCCG |
| Sequence-based reagent | Internal rev sacB (9C addition) | IDT | ODP596 | GTTGGGGGGGGGCATCGTTCATGTCTCCTTTTTTATG |
| Sequence-based reagent | Internal fw sacB (9C addition) | IDT | ODP597 | ATGCCCCCCCCCAACATCAAAAAGTTTGCAAAACAAG |
| Sequence-based reagent | rev sacB | IDT | ODP598 | GATAAGCTTCGAATTACTATCAATAAGTTGGAGTCATTACC |
| Sequence-based reagent | Internal rev sacB (6C addition) | IDT | ODP614 | GTTGGGGGGCATCGTTCATGTCTCCTTTTTTATG |
| Sequence-based reagent | Internal fw sacB (6C addition) | IDT | ODP615 | ATGCCCCCCAACATCAAAAAGTTTGCAAAACAAG |
| Sequence-based reagent | fw PCR screening and seq pmsg419 cloning | IDT | ODP236 | CTCCCTATCAGTGATAGATAGGCTCTGG |
| Sequence-based reagent | rev PCR screening and seq pmsg419 cloning | IDT | ODP237 | CATGACCAACTTCGATAACGTTCTCGG |
| Sequence-based reagent | fw PCR screening and seq pDB60 cloning | IDT | ODP474 | TGATTCTGTGGATAACCGTATTACCGCCTTTG |
| Sequence-based reagent | rev PCR screening and seq pDB60 cloning | IDT | ODP475 | AAGGCCCAGTCTTTCGACTGAGC |
| Sequence-based reagent | fw rpoB PCR | IDT | ODP378 | CAAGAAGCTGGGCCTGAACGC |
| Sequence-based reagent | rev rpoB PCR | IDT | ODP379 | GCGGTTGGCGTCGTCGTG |
| Sequence-based reagent | rpoB seq | IDT | ODP380 | GAGCGTGTCGTGCGTGAG |
| Sequence-based reagent | fw kan or sacB PCR | IDT | ODP476 | TGGCCTTTTGCTGGCCTTTTGC |
| Sequence-based reagent | rev kan PCR | IDT | ODP477 | TTCAACAAAGCCGCCGTCCC |
| Sequence-based reagent | kan seq | IDT | ODP479 | ACTGAATCCGGTGAGAATGG |
| Sequence-based reagent | rev sacB PCR | IDT | ODP172 | TTAGACGTAATGCCGTCAATCGTC |
| Sequence-based reagent | sacB seq | IDT | ODP474 | TGATTCTGTGGATAACCGTATTACCGCCTTTG |
| Sequence-based reagent | 5' 32P-labeled primer DNA strand | IDT | SG-FS1 | CGTGTCGCCCTTC |
| Sequence-based reagent | Unlabeled template strand (4T) | IDT | SG-FS1 | GGGTTTTGAAGGGCGACACG |
| Sequence-based reagent | Unlabeled template strand (6T) | IDT | SG-FS1 | GGGTTTTTTGAAGGGCGACACG |
| Sequence-based reagent | Unlabeled template strand (8T) | IDT | SG-FS1 | GGGTTTTTTTTGAAGGGCGACACG |
| Sequence-based reagent | Unlabeled template strand (4A) | IDT | SG-FS1 | CCCAAAAGAAGGGCGACAC |
| Sequence-based reagent | Unlabeled template strand (6A) | IDT | SG-FS1 | CCCAAAAAAGAAGGGCGACAC |
| Sequence-based reagent | Unlabeled template strand (8A) | IDT | SG-FS1 | CCCAAAAAAAAGAAGGGCGACAC |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83094/elife-83094-mdarchecklist1-v1.docx
-
Source data 1
Primary data for all non-gel data elements in the figures and figure supplements.
- https://cdn.elifesciences.org/articles/83094/elife-83094-data1-v1.xlsx





