Why did glutamate, GABA, and melatonin become intercellular signalling molecules in plants?
Figures
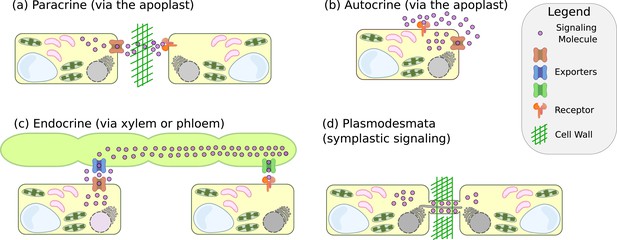
Modes of intercellular signalling.
(a) Paracrine signalling, where cell-to-cell communication is mediated through the apoplastic space. (b) Autocrine signalling - similar to paracrine signalling, but when the sender cell also acts as a receiver cell. (c) Long-distance endocrine signalling that is mediated by translocation of the signalling molecules through the vascular tissue of plants. (d) Direct cell-to-cell communication and signalling through the plasmodesmata.
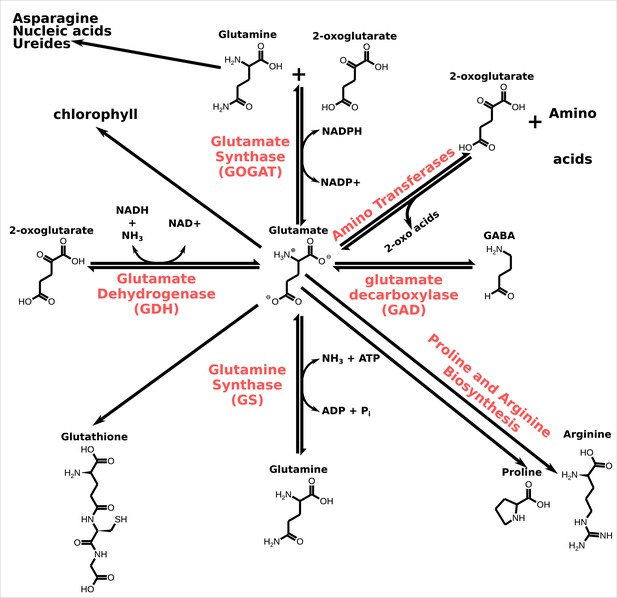
Glutamate metabolism.
In red, the names of the enzymes that catalyse the chemical conversion. Similar reactions in the glutamate metabolic network also occur in animals (see Schousboe et al., 2014). Note that the figure does not show the balance of the chemical reactions.
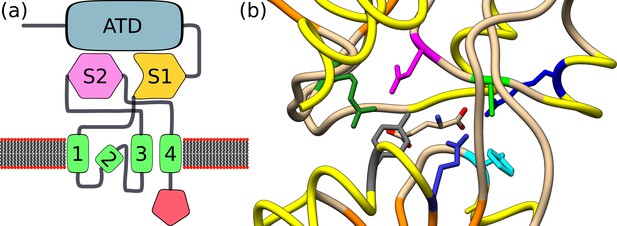
Glutamate receptor-like (GLRs), their agonists, and animal ionotropic glutamate receptors.
(a) General structure of the ionotropic glutamate receptor. An ionotropic glutamate receptor (iGluR) monomer is composed of an amino-terminal domain (ATD) followed by the first half of the ligand-binding domain (S1). S1 is connected to three transmembrane segments (shown in green and numbered 1–3). The second transmembrane segment spans the membrane only partly. The Third transmembrane segment is connected to the second half of the ligand-binding domain. Finally, a fourth transmembrane segment is connected to the intracellular C-terminal domain. The figure is based on De Bortoli et al., 2016. Plant GLRs show a similar structure with a highly diverse C-terminal domain, three membrane-spanning segments (plus one partly spanning the membrane), and the divided ligand-binding domain. However, clade III ATD shows more homology to members of the sub-family C of G-protein-coupled receptors (GPCRs), which contain metabotropic glutamate receptors (mGluRs) and γ-aminobutyric acid B (GABAB) receptors in animals (Wudick et al., 2018a). (b) Crystal structure of the binding pocket of Arabidopsis GLR3.3 with glutamate as a ligand-based on Alfieri et al., 2020 (Protein Data Bank code: 6R85). Glutamate is depicted in sticks. Residues that were implicated in binding glutamate are Phe133 in cyan, Arg88 inbBlue, Ala83 in green, Arg11 in forest green, Tyr180 in grey, Glu177 in medium blue, and Asp81 in magenta. Backbone – in yellow α-helices, in orange β-strands.
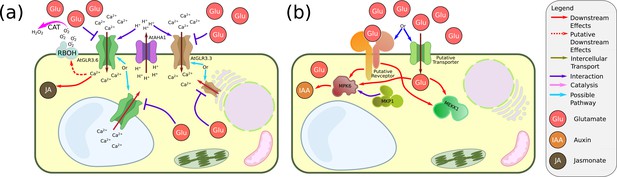
Glutamate signalling in plants.
(a) Glutamate signalling via glutamate receptor-likes (GLRs). It was suggested that, following wounding, glutamate is secreted from the phloem. In parallel, the ATPase AtAHA1 is deactivated, and the apoplast pH rises. These processes activate AtGLR3.3 or AtGLR3.6 (or both) and cause electric potential propagation (not shown) as well as Ca2+ influx to the cytoplasm. The activation of AtGLR3.3 and AtGLR3.6 initiates a signalling cascade that results in jasmonate synthesis. In some plants (e.g. tomato), it might also lead to H2O2 signalling via RESPIRATORY BURST OXIDASE HOMOLOG (RBOH). Note that AtGLR3.3 and AtGLR3.6 are not localised to the same cell populations in the leaves (phloem sieve elements and xylem contact cells, respectively). It was also found that they might not localise to the plasma membrane (ER and vacuole, respectively). (b) Glutamate in environmental-root tip signalling. Putative glutamate signalling during RSA restructuring. A putative receptor or a putative transporter (not identified yet) transduces the presence of extracellular glutamate to the cytoplasm. Downstream, both MEKK1 and the pair MKP1/MPK6 are influenced. Even downstream from MKP1/MPK6, the level of auxin is modified.
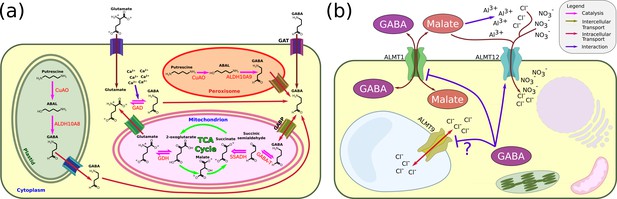
GABA in plant physiology and signalling.
(a) GABA synthesis (based on Fait et al., 2008 and Gilliham and Tyerman, 2016). GABA is synthesised via two main routes. In the context of the GABA shunt, it is synthesised in the cytoplasm from glutamate by glutamate decarboxylase (GAD) and is then imported to the mitochondria, where it is converted to succinic semialdehyde. In the plastids and the peroxisomes, it is synthesised from the diamine putrescine. The ability of the GABA shunt to control the C/N ratio makes GABA a pivotal metabolite under various stress conditions. Similarly, the importance of polyamines in stress mechanisms also connects GABA to the maintenance of cell physiology. In addition, since the GAD reaction consumes a proton, GABA might act as a pH regulator. (b) GABA in signalling. Suggested GABA signalling events depend on the ALMT protein family. In the roots, GABA inhibits the transport of malate through ALMT1 upon Al3+ stress. In the leaves, GABA might regulate stomatal closure during drought by inhibiting Cl− transport through the tonoplast localised ALMT9 (which does not count as an intercellular signalling event by our criteria). GABA can also regulate stomatal closure through ALMT12. However, the precise physiological relevance of this process is still not clear. GABA inhibition of ALMT1 malate transport might be achieved through an interaction with a peptide moiety at the cytoplasmic side. It is not known whether this is achieved primarily by GABA imported to the cell via ALMT1 itself or by endogenous cytoplasmic GABA. The possible involvement of GABA in pollen tube growth is not shown since the underlying mechanism is not yet deciphered. Similarly, the putative interactions between GABA and GORK is not shown. GAD, glutamate decarboxylase; SSR, semialdehyde reductase; GABA-T, GABA transaminase; SSADH, succinic semialdehyde dehydrogenase; GABP, mitochondrial GABA transporter; GAT, GABA transporters; CuAO, copper-containing amine oxidase; ALDH, aldehyde dehydrogenase.
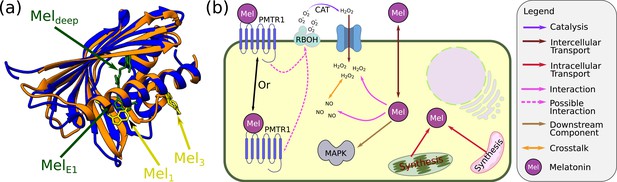
Melatonin signalling in plants.
(a) Crystal structure of the PR-10 proteins that bind melatonin. Blue: Hyp-1 from St. John’s wort based on Protein data bank #5I8F (Sliwiak et al., 2016). Hyp-1 binds 2–3 melatonin molecules (the two with the more sound evidence are shown). Orange: LLPR-10.2B from yellow lupin with the two sites where melatonin binds the protein (based on PDB #5MXB; Sliwiak et al., 2018). (b) Signalling by melatonin. Due to its amphipathic properties, exogenous melatonin can shuttle between the apoplast and the symplast spaces. In both spaces, it can scavenge reactive oxygen species (ROS) and mediate ROS signalling. In the symplastic space, melatonin can also interact with reactive nitrogen species (RNS) and, in particular, nitric oxide, thereby mediating downstream NO signalling. In addition, it can interact with PMTR1, which is either a membrane or a cytoplasmic protein (still under dispute). PMTR1 can activate RBOH and initiate ROS signalling. In addition, melatonin probably works via other mechanisms that include a downstream MAPK signalling cascade.
Tables
Main indication of the first triad function in plant signalling and the second triad function in plant physiology.
Yellow background: molecules from the first triad that act as plant signalling (and particularly intercellular) molecules and the best examples for their participation in signalling processes. Green background: molecules from the second triad where there is currently a lack of evidence of their participation in intercellular signalling in plants and examples of processes where they do participate. References related to the figure are glutamate (Kwaaitaal et al., 2011; Li et al., 2019; López-Bucio et al., 2018; Mousavi et al., 2013; Nguyen et al., 2018; Toyota et al., 2018; Walch-Liu et al., 2006; Wang et al., 2019a; Yoshida et al., 2016); GABA (AlQuraan, 2015; Guo et al., 2020; Li et al., 2016a; Malekzadeh et al., 2013; Mustroph et al., 2014; Palanivelu et al., 2003; Ramesh et al., 2015; Seifikalhor et al., 2020; Wang et al., 2014; Wang et al., 2017a; Xu et al., 2021a); melatonin (Chen et al., 2019; Chen et al., 2018; Chen et al., 2017; Erland et al., 2018; Giridhar and Ravishankar, 2009; Li et al., 2014a; Liang et al., 2017b; Pelagio-Flores et al., 2012; Tan et al., 2014; Wei et al., 2018; Zhang et al., 2019; Zhu et al., 2019); serotonin (He et al., 2021; Kang et al., 2009; Mukherjee et al., 2014; Pelagio-Flores et al., 2016; Pelagio-Flores et al., 2011; Shukla et al., 2021; Wan et al., 2018a ; Wan et al., 2018b); dopamine (Dai et al., 1993; Gomes et al., 2014; Guidotti et al., 2013; Protacio et al., 1992; Stefi et al., 2019); and acetylcholine (Bamel et al., 2015; Bamel et al., 2007; Bamel and Gupta, 2018; Di Sansebastiano et al., 2014; Erland and Saxena, 2019; Qin et al., 2020; Tezuka et al., 2007). Note that the reference list is not exhaustive. For full references, see the main text. Also, for full details of the effects related to each molecule, see the full text.
| Molecule | Effect | ||
|---|---|---|---|
| Processes indicative of signalling/participation in physiology | Putative receptor | ||
| Signalling | Glutamate |
| GRL3.3 GRL3.6 SlGLR3.5 |
| GABA |
| ALMT1 ALMT9 ALMT12 | |
| Melatonin |
| PMTR1 RD-10 | |
| Non-signalling | Serotonin |
| – |
| Dopamine |
| – | |
| Acetylcholine |
| – | |
Summary of the effects of different molecules discussed in this article on the plant root system.
For the exogenous glutamate effect, see Figure 2 in Walch-Liu et al., 2006. For the exogenous GABA effect, see Figure 1 in Barbosa et al., 2010, Figure 2 in Xie et al., 2020, and Ramesh et al., 2015. For exogenous treatment with melatonin, see Figure 2 and Figure 5 in Pelagio-Flores et al., 2012, Figure 1 in Bajwa et al., 2014, Figure 1 in Liang et al., 2017b, and Erland et al., 2018. For the exogenous serotonin effect, see Figure 2 in Pelagio-Flores et al., 2011, Figure 1 in Pelagio-Flores et al., 2016, Figure 1 in Wan et al., 2018b, and Figure 1 in Wan et al., 2018a. For exogenous dopamine effect, see Figure 3 in Liang et al., 2017a. For the exogenous acetylcholine effect, see Figure 2 in Sugiyama and Tezuka, 2011, Figures 5 and 6 in Murata et al., 2015, and Figure 1 in Bamel et al., 2007. Note that the reference list is not exhaustive. For full references, see the main text. Conc: concentration. Yellow background: metabolites that are considered to be intercellular signalling molecules by our criterion. Green background: metabolites that are not considered to be intercellular signalling molecules by our criterion.
| Molecule | Effect |
|---|---|
| Effect on the root system architecture | |
| Glutamate |
|
| GABA |
|
| Melatonin |
|
| Serotonin |
|
| Dopamine |
|
| Acetylcholine |
|





