Rapid cell type-specific nascent proteome labeling in Drosophila
Figures
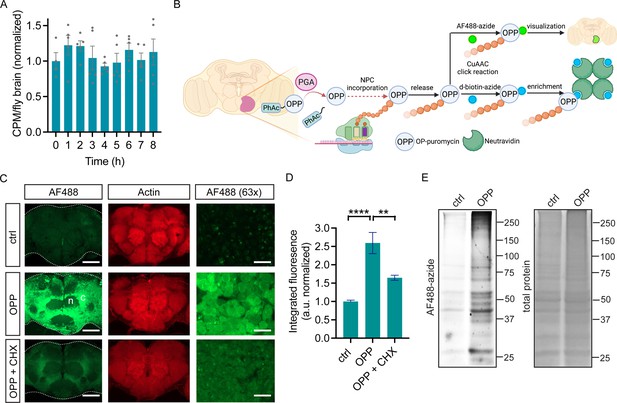
O-propargyl-puromycin (OPP) labeling in Drosophila brain.
(A) Global protein synthesis (35S-met/cys incorporation) is unaltered for 8 hr in newly-isolated w1118 whole brain preparations (ANOVA, n.s. n=4–5 groups of 8–10 brains per timepoint). (B) Schematic illustrating cell type-specific protein synthesis labeling by PGA-dependent OPP incorporation (POPPi) for visualization or capture of the nascent proteome. Spatially targeted penicillin G acylase (PGA) expression catalyzes phenylacetyl-OPP (PhAc-OPP) blocking group removal, liberating OPP for incorporation into nascent polypeptide chains (NPC). OP-puromycylated proteins can be visualized by confocal microscopy following conjugation to a fluorescent-azide or enriched following conjugation to desthiobiotin-azide. (C) Newly-synthesized protein (from w1118 brains) visualized by AF488-azide after OPP incubation but not without OPP (ctrl) and diminished signal with cycloheximide (CHX). Labeling appears stronger in cell bodies within the cell cortex (C) than in the neuropil (n). Higher magnification (63 x) images are from the cell cortex. Scale bars are 60 μM (or 10 μM for 63 x). (D) Quantitation of (C) revealing a significant effect of the treatment group (ANOVA, p<0.0001, Bonferroni post-test, **p<0.01, ****p<0.0001, n=8–11 brains/group). (E) In-gel fluorescence of brain protein extracts, ctrl is no OPP. Data are mean ± SEM. Schematic in B created on Biorender. See also Figure 1—figure supplements 1 and 2.
-
Figure 1—source data 1
Source gel images for Figure 1E.
- https://cdn.elifesciences.org/articles/83545/elife-83545-fig1-data1-v1.zip
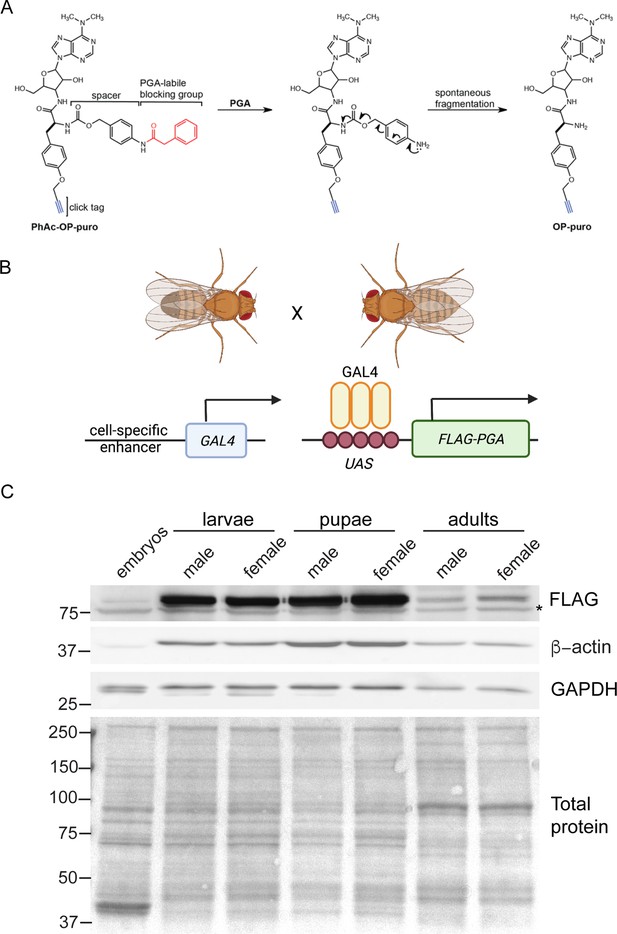
Penicillin G acylase (PGA)-dependent conversion of phenylacetyl-OPP (PhAc-OPP) to O-propargyl-puromycin (OPP) in Drosophila.
(A) Scheme illustrating the conversion of PhAc-OPP to OPP by penicillin G acylase (PGA). PGA catalyzes the removal of the phenylacetyl blocking group and the carbamate spacer then undergoes spontaneous fragmentation to generate OPP. (B) Scheme illustrating expression of N-terminally FLAG-tagged PGA under a cell-specific driver of choice using the binary GAL4/UAS system. (C) PGA expression levels within whole embryos or the brains of L3 larvae, pupae or adult flies expressing PGA via the ubiquitous Actin5C-GAL4 driver. Asterisk, non-specific band. Both loading controls (β-actin and GAPDH) are divergent in expression, size, and/or number of bands between embryos, larvae, pupae, and adults, hence a Ponceau stain is shown for protein loading. Illustration in B created on Biorender.
© 2016, American Chemical Society. Panel A is reprinted (adapted) with permission from Scheme 1 of Barrett et al., 2016. It is not covered by the CC-BY 4.0 license and further reproduction of this panel would need permission from the copyright holder.
-
Figure 1—figure supplement 1—source data 1
Source western blots for Figure 1—figure supplement 1C.
- https://cdn.elifesciences.org/articles/83545/elife-83545-fig1-figsupp1-data1-v1.zip
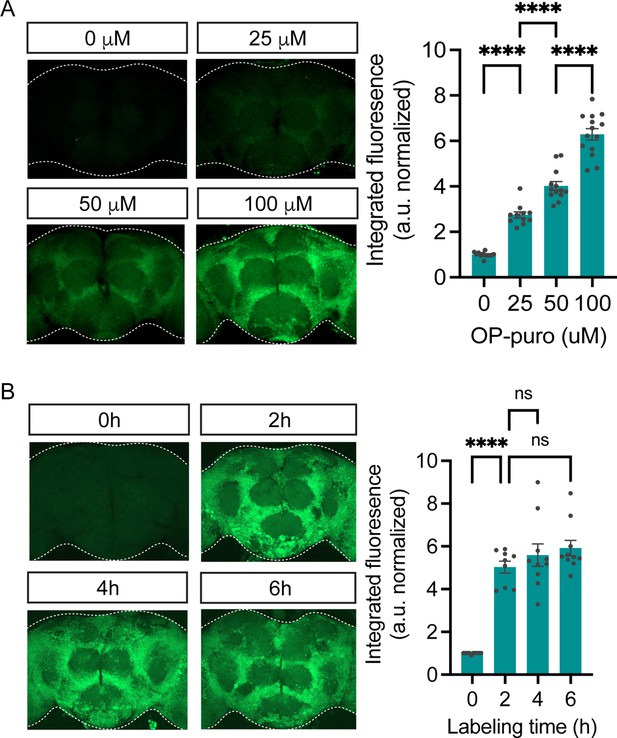
O-propargyl-puromycin (OPP) nascent proteome labeling is concentration and time-dependent.
(A) Significant effect of OPP concentration on OPP labeling in standard laboratory control flies (w1118, ANOVA, p<0.0001, Bonferroni post-tests, ****p<0.0001, n=11–14 brains/group). Labeling time was 2 hr. (B) Significant effect of labeling time on OPP labeling in w1118 flies (ANOVA, Bonferroni post-tests, ****p<0.0001, n=9–10 brains/group). 50 μM OPP was used. Data are mean ± SEM.
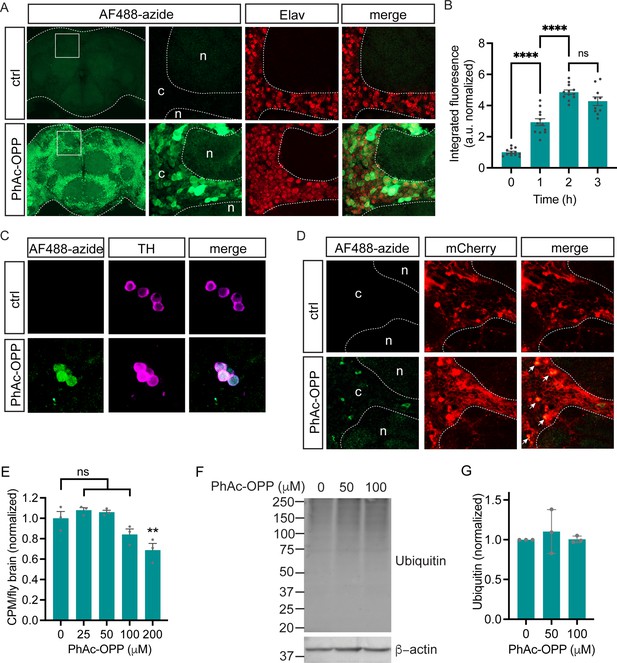
Cell type-specific protein synthesis labeling with phenylacetyl-OPP (PhAc-OPP).
(A) Brains from elavC155-GAL4;UAS-PGA flies incubated with PhAc-OPP show widespread protein synthesis labeling in neurons, labeled with Elav neuronal nucleus marker. No labeling in vehicle-treated brains (ctrl). Labeling appears highest in the cell bodies of the cell cortex (c) and minimal in the neuropil (n). (B) AF488-azide quantitation after varying durations of PhAc-OPP labeling in pan-neuronal penicillin G acylase (PGA) expressing flies. Significant effect of PhAc-OPP incubation time on labeling (ANOVA, Bonferroni post-test, ****p<0.0001, n=10–12 brains per group). (C) Protein synthesis labeling in isolated brains from flies expressing PGA in dopamine neurons (TH-GAL4/UAS-PGA). TH, tyrosine hydroxylase. (D) Protein synthesis labeling following pan-glial expression of PGA and a membrane-tethered mCherry reporter (Repo-GAL4, UAS-mCD8::mCherry/UAS-PGA). Glial cell bodies and neuron-encapsulating surface areas are mCherry-positive. Arrows indicate glial cell bodies positive for both mCherry and AF488, indicative of glial protein synthesis labeling. Controls in A, C, and D are vehicle-treated brains. (E) Significant effect of 200 μM PhAc-OPP on global protein synthesis in flies expressing PGA ubiquitously via Actin5C-GAL4 (ANOVA, Bonferroni post-test, **p<0.01, n=3 groups of 8–10 brains/group). (F) No significant effect of PhAc-OPP on total protein ubiquitination in flies expressing PGA ubiquitously, quantified in G (ANOVA, n=3 groups of 20 brains/group). Data are mean ± SEM. See also Figure 2—figure supplement 1.
-
Figure 2—source data 1
Source western blots for Figure 2F.
- https://cdn.elifesciences.org/articles/83545/elife-83545-fig2-data1-v1.zip
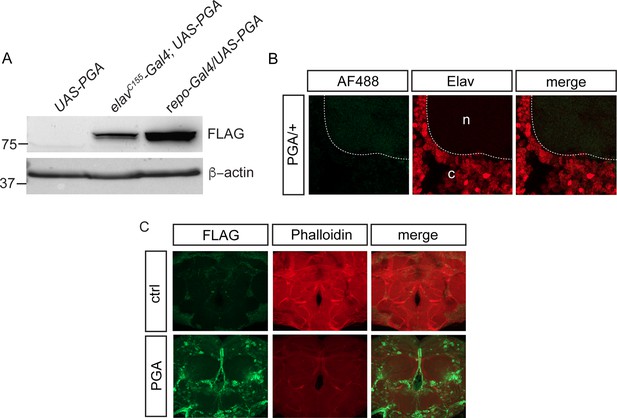
Cell-specific labeling via penicillin G acylase (PGA) expression.
(A) FLAG-PGA expression via elavC155-GAL4 or repo-GAL4 in whole brain extracts. (B) No phenylacetyl-OPP (PhAC-OPP) labeling in UAS-PGA/+ control fly brains lacking the elavC155-GAL4 driver, incubated with PhAc-OPP. c, cell cortex showing positive immunostaining for the neuronal nuclear protein Elav. n, neuropil. (C) FLAG (L5 clone) immunostaining for PGA expression pattern in elavC155-GAL4>UAS PGA fly brain (PGA). Control is elavC155-GAL4/+.
-
Figure 2—figure supplement 1—source data 1
Source western blots for Figure 2—figure supplement 1A.
- https://cdn.elifesciences.org/articles/83545/elife-83545-fig2-figsupp1-data1-v1.zip
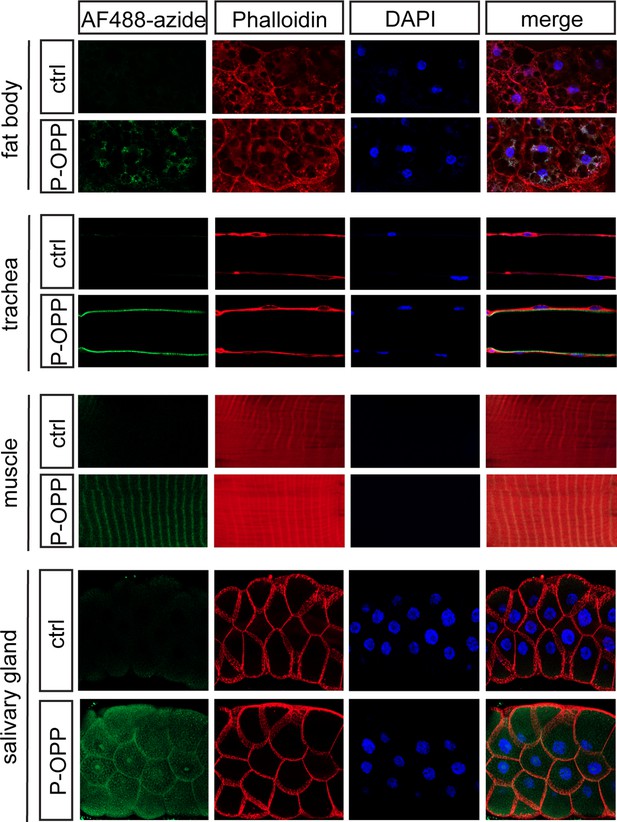
Protein synthesis labeling in other major tissues using phenylacetyl-OPP (PhAc-OPP).
Tissues from L3 larvae incubated with PhAc-OPP show widespread protein synthesis labeling, visualized following O-propargyl-puromycin (OPP) conjugation to AF488-azide. All larvae expressed penicillin G acylase (PGA) via Actin5C-Gal4.
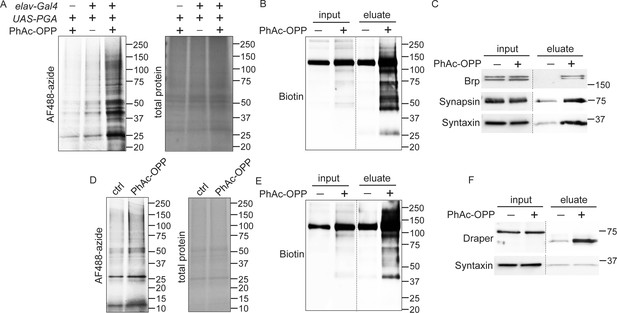
Capture and identification of newly-synthesized proteins from neurons and glial.
(A) In-gel AF488 fluorescence across a broad range of proteins in fly brain extracts expressing penicillin G acylase (PGA) pan-neuronally and following incubation with phenylacetyl-OPP (PhAc-OPP). Controls groups are no PGA expression (with PhAc-OPP) or no PhAc-OPP (with PGA expression). (B) Detection of total desthiobiotin-tagged protein in brain lysates (input) or pulldown fractions (eluate) following pan-neuronal PGA expression, PhAc-OPP incubation, conjugation of OPP-labeled protein to desthiobiotin azide and neutravidin bead pulldown. A strong background band of unknown endogenously biotinylated protein is observed between 100–150 kDa. (C) Enrichment of neuronal proteins Brp, Synapsin, and Syntaxin following neutravidin pulldown of desthiobiotin-tagged protein from pan-neuronal elav-GAL4/UAS-PGA brain extracts. (D) Pan-glial PGA expression (Repo-GAL4/UAS-PGA) promotes broad O-propargyl-puromycin (OPP) protein labeling in brain extracts following PhAc-OPP incubation but not vehicle control. Enrichment of total desthiobiotin-tagged protein seen following neutravidin bead pulldown of the glial proteome, detected with anti-biotin (D) and enrichment of the glial-specific protein Draper (with apparent mol. weight shift between input and eluate) from Repo-GAL4/UAS-PGA extracts, but not the neuron-specific protein Syntaxin. Data are representative of three independent experiments.
-
Figure 4—source data 1
Source western blots for Figure 4.
- https://cdn.elifesciences.org/articles/83545/elife-83545-fig4-data1-v1.zip
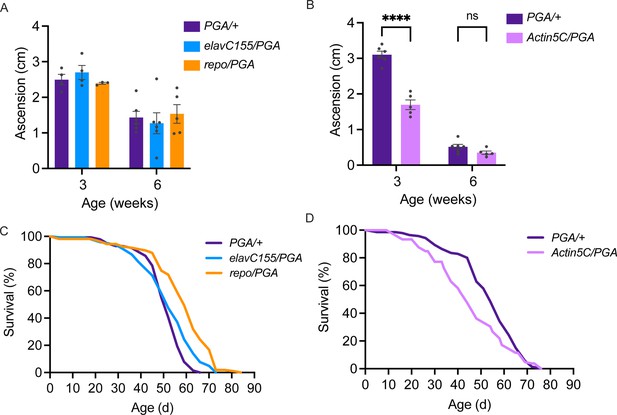
No deleterious effect of cell-specific penicillin G acylase (PGA) expression.
(A) No significant effect of pan-neuronal or pan-glial PGA expression on negative geotaxis behavior at 3 and 6 weeks of age (two-way ANOVA for the effect of age (p<0.0001) and genotype, (n.s.), n=3–6 groups of 25 flies/genotype/age). (B) Significant effect of ubiquitous PGA expression on negative geotaxis behavior at 3 weeks of age (two-way ANOVA for the effect of age (p<0.0001) and genotype (p<0.0001), Bonferroni post-test, ****p<0.0001, n=5–6 groups of 25 flies/genotype/age). (C) Survival is slightly extended upon pan-neuronal or pan-glial PGA expression. (D) Significant effect of ubiquitous PGA expression on survival. For C and D, see Tables 1 and 2 for experimental n, lifespan metrics, and log-rank (Mantel-Cox) comparison results. Data are mean ± SEM. See also Figure 5—figure supplement 1.
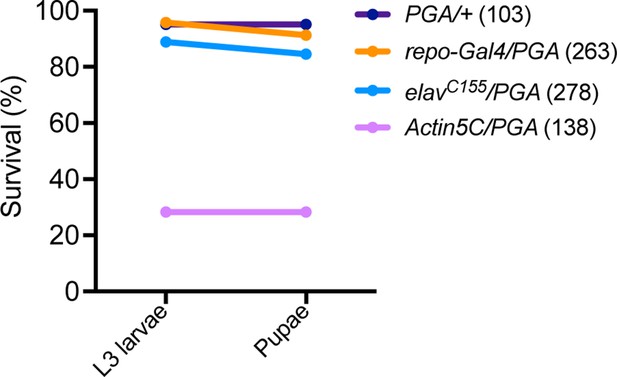
Larval lethality following ubiquitous penicillin G acylase (PGA) expression.
Numbers in parentheses are the number of flies tested.
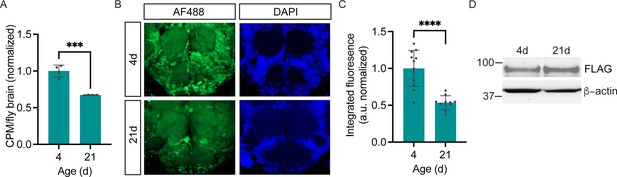
Age-dependent decline in neuronal protein synthesis rate.
(A) Significant age-related decline in whole brain protein synthesis in aging flies, measured by 35S-met/cys labeling (Student’s t-test, ***p<0.001, n=4 groups of eight brains/age). (B) Nascent proteome labeling in young (4-day-old) vs. aged (21-day-old) fly brains expressing pan-neuronal penicillin G acylase (PGA) following phenylacetyl-OPP (PhAc-OPP) incubation. (C) The significant effect of aging on protein synthesis (Student’s t-test, p<0.0001, n=10–12 brains/group). Fluorescence signals at each age was derived by subtracting from the mean of a no-label control brain population tested in parallel. (D) PGA expression is comparable in 4- and 21-day-old elavC155-GAL4/UAS-PGA fly brains. Data are mean ± SEM.
-
Figure 6—source data 1
Source western blots for Figure 6D.
- https://cdn.elifesciences.org/articles/83545/elife-83545-fig6-data1-v1.zip
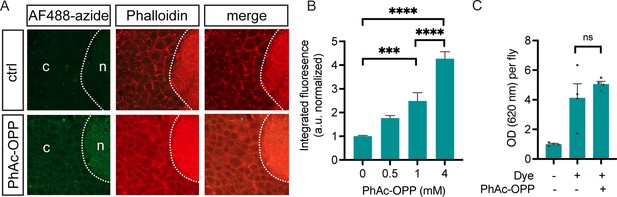
Protein synthesis labeling via dietary phenylacetyl-OPP (PhAc-OPP) administration.
(A) Subtle detection of protein synthesis labeling by AF488-azide following exposure of pan-neuronal penicillin G acylase (PGA)-expressing flies to dietary PhAc-OPP (4 mM) for 48 hr. Brains were counterstained with Dylight Phalloidin-650. (B) AF488-azide quantitation after varying concentrations of PhAc-OPP exposure in pan-neuronal PGA-expressing flies. Significant effect of PhAc-OPP concentration on labeling (ANOVA, Bonferroni post-test, ***p<0.001, ****p<0.0001, n=12–17 brains per group). (C) No significant effect of PhAc-OPP (4 mM) on food intake levels over 4 hr (ANOVA, Bonferroni post-test, ns, n=4 groups of eight flies per condition).
Tables
Ubiquitous penicillin G acylase (PGA) expression effect on survival.
| Genotype | N | Median lifespan (d) | Mean lifespan (d) | Log-rank (vs PGA/+ctrl) |
|---|---|---|---|---|
| UAS-PGA/+ | 142 | 54 | 53.1 | - |
| Actin5C-GAL4/UAS-PGA | 110 | 44 | 45.6 | .006 |
Neuronal and glial penicillin G acylase (PGA) expression effect on survival.
| Genotype | N | Median lifespan (d) | Mean lifespan (d) | Log-rank (vs. PGA/+ctrl) |
|---|---|---|---|---|
| UAS-PGA/+ | 152 | 52 | 50.8 | - |
| elavC155-GAL4/UAS-PGA | 147 | 52 | 52.1 | 0.002 |
| Repo-GAL4/UAS-PGA | 112 | 63 | 58.4 | <0.001 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | UAS-PGA | This paper | FLAG-tagged penicillin G acylase under UAS control | |
| Genetic reagent (D. melanogaster) | Actin5C-Gal4 | Bloomington Drosophila Stock Center | BDSC:25374 FLYB:FBti012 7834; RRID:BDSC_ 25374 | FlyBase symbol: P{Act5C-GAL4-w}E1 |
| Genetic reagent (D. melanogaster) | Elav(C155)-Gal4 | Bloomington Drosophila Stock Center | BDSC:458 FLYB:FBti000 2575; RRID:BDSC_ 458 | FlyBase symbol: P{GawB}elav [C155] |
| Genetic reagent (D. melanogaster) | Repo-Gal4 | Bloomington Drosophila Stock Center | BDSC:7415 FLYB:FBti001 8692 RRID:BDSC_ 7415 | FlyBase symbol: P{GAL4}repo |
| Genetic reagent (D. melanogaster) | TH-Gal4 | Bloomington Drosophila Stock Center | BDSC:8848 FLYB:FBti007 2936 RRID:BDSC_ 8848 | FlyBase symbol: P{ple-GAL4.F}3 |
| Biological sample (D. melanogaster) | Drosophila brain explants | This paper | Freshly isolated from various D. melanogaster genotypes | |
| Antibody | Anti-Biotin rabbit polyclonal | Bethyl Laboratories Inc. | Cat: A150109A | 1:1000 |
| Antibody | Anti-Brp (nc82) mouse monoclonal | DSHB | Cat: nc82 | 1:50 |
| Antibody | Anti-Synapsin (3C11) mouse monoclonal | DSHB | Cat: 3C11 | 1:500 |
| Antibody | Anti-Syntaxin (8C3) mouse monoclonal | DSHB | Cat: 8C3 | 1:500 |
| Antibody | Anti-Draper 8A1 mouse monoclonal | DSHB | Cat: 8A1 | 1:400 |
| Antibody | Anti-Draper 5D14 mouse monoclonal | DSHB | Cat: 5D14 | 1:400 |
| Antibody | Anti-Elav 9F8A9 mouse monoclonal | DSHB | Cat: 9F8A9 | 1:100 |
| Antibody | Anti-FLAG (M2) mouse monoclonal | Millipore Sigma | Cat: F1804 | 1:500 |
| Antibody | Anti-FLAG (L5) mouse monoclonal | Novus Biologicals | Cat: NBP1-06712 | 1:1000 |
| Antibody | Anti-Actin-HRP (AC15) mouse monoclonal | Millipore Sigma | Cat: A3854 | 1:2000 |
| Antibody | Anti-GAPDH (GA1R) mouse monoclonal | ThermoFisher | Cat:MA5-15738 | 1:10,000 |
| Antibody | Anti-TH mouse monoclonal | Immunostar | Cat: 22941 | 1:1000 |
| Antibody | Anti-Ubiquitin (P4D1) rabbit monoclonal | Cell Signaling Technology | Cat: 3936 | 1:1000 |
| Commercial assay or kit | Click-iT Plus OPP Alexa Fluor 488 Protein Synthesis Assay Kit | ThermoFisher | Cat: C10456 | |
| Chemical compound, drug | PhAc-OPP | This paper and reference (Barrett et al., 2016) | See reference (Barrett et al., 2016) for chemical synthesis details | |
| Chemical compound, drug | Desthiobiotin azide | Click Chemistry Tools | Cat: 50-210-7822 | |
| Chemical compound, drug | TBTA (Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine) | Millipore Sigma | Cat:678937 | |
| Chemical compound, drug | Cu(I)Br | Millipore Sigma | Cat: 61163 | |
| Chemical compound, drug | Neutravidin agarose | Thermo Scientific | Cat: P129204 | |
| Chemical compound, drug | Colloidal blue staining kit | ThermoFisher | Cat: LC6025 |





