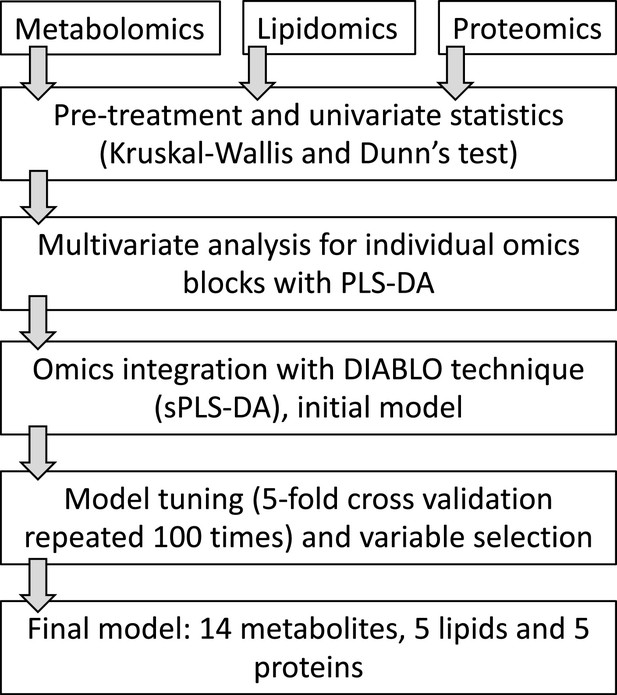
The ‘ForensOMICS’ approach for postmortem interval estimation from human bone by integrating metabolomics, lipidomics, and proteomics
Figures
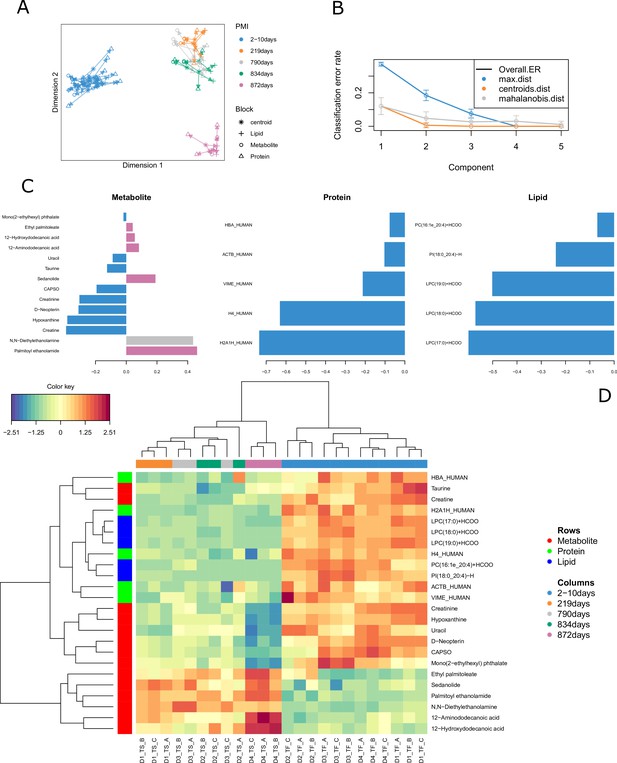
Results for the tuned model.
(A) Arrow plot showing multiblock contexts for the overall model. (B) Optimal number of components to explain model variable calculated via cross-validation (error bars provide standard deviation). (C) Loading plot showing how each variable contributes to the covariance of each group. (D) The clustered image map (CIM) shows the selected compounds in the final model. It is possible to see that most compounds decrease in intensity after decomposition except for few metabolites and two lipids that specifically increase in certain postmortem interval (PMI) intervals.
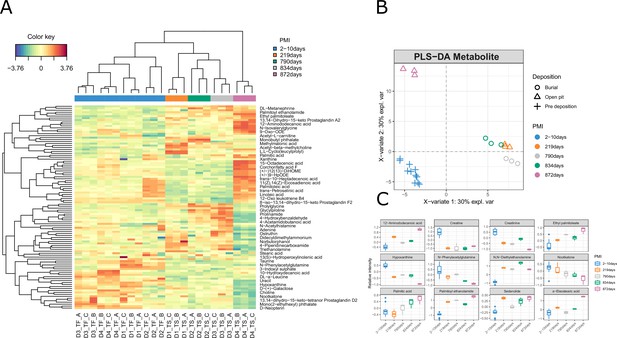
Results for the metabolomics data.
(A) Clustered image map (CIM), (B) sample plot, (C) and boxplot for the metabolomics data.
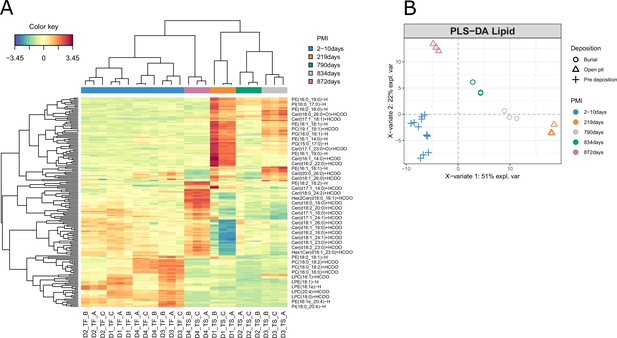
Results for the lipidomics data.
(A) Clustered image map (CIM) and (B) sample plot for the lipidomics data.
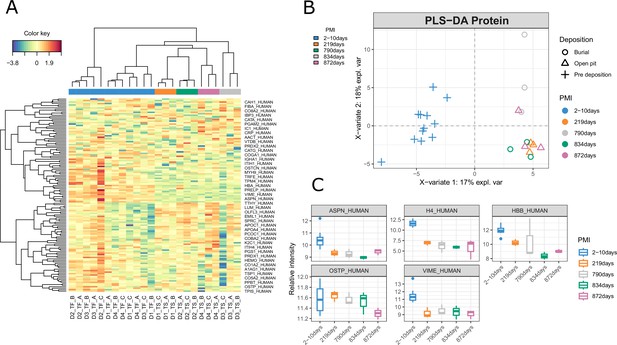
Results for the proteomics data.
(A) Clustered image map (CIM), (B) sample plot, (C) and boxplot for the proteomics data.
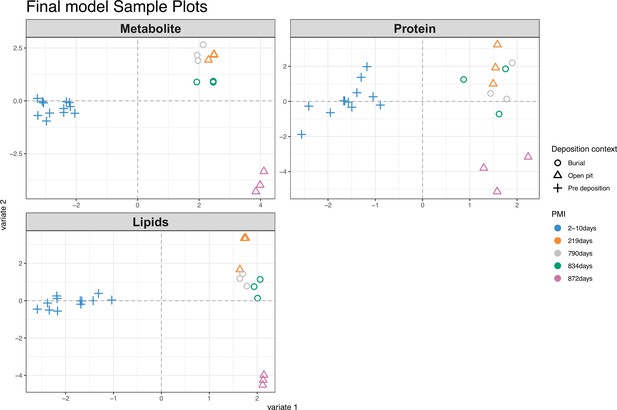
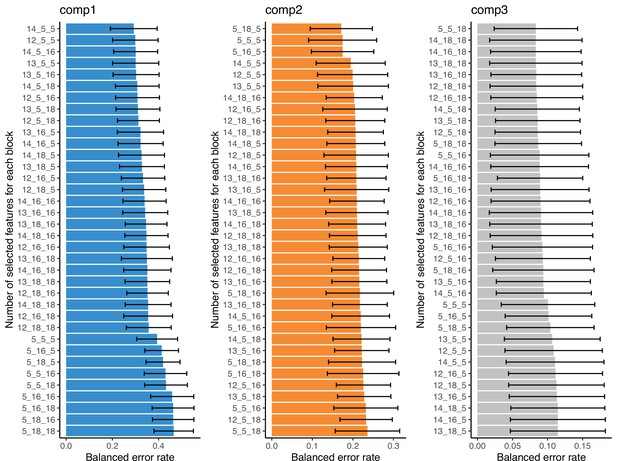
Score plots for partial least square discriminant analysis (PLS-DA) results of all the omics blocks considered.
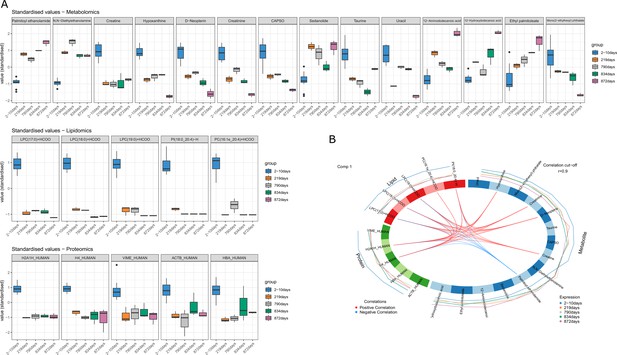
DIABLO selected variables correlated with PMI.
(A) Boxplots of the selected variables after tuning that shows variation with postmortem interval (PMI). Variables are expressed in standardized values. (B) Correlation between different omics blocks highlighting the correlations between different compounds obtained with the three omics selected in the final discriminant analysis model.
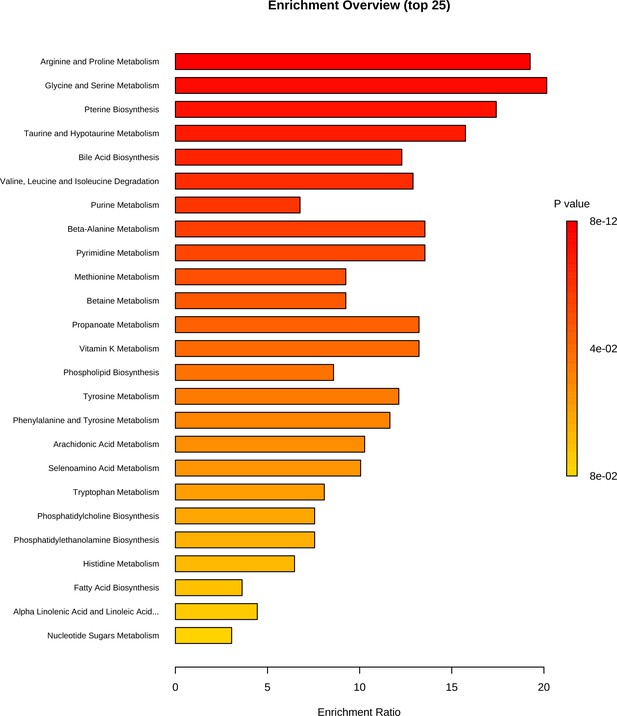
Metabolite set enrichment analysis based on differentially expressed metabolites identified in bone.

Positioning of the bodies in the single graves (left) pre-decomposition and (right) after complete skeletonization.
Tables
Sample composition, demographics, deposition context, and postmortem interval (PMI).
The sample ID column reports the biological replicates used. Additional information on the body donors and observations made during collection of bone samples (e.g., medical treatments, bone colour, and density) can be found in the supplementary information in Mickleburgh et al., 2021.
| Sample ID | Sex | Age (years) | PMI | Deposition context | |
|---|---|---|---|---|---|
| Pre-deposition samples | |||||
| D1_TF_A | Female | 91 | 10 days | Open pit | |
| D1_TF_B | Female | 91 | 10 days | Open pit | |
| D1_TF_C | Female | 91 | 10 days | Open pit | |
| D2_TF_A | Female | 67 | 2 days | Burial | |
| D2_TF_B | Female | 67 | 2 days | Burial | |
| D2_TF_C | Female | 67 | 2 days | Burial | |
| D3_TF_A | Female | 61 | 3 days | Burial | |
| D3_TF_B | Female | 61 | 3 days | Burial | |
| D3_TF_C | Female | 61 | 3 days | Burial | |
| D4_TF_A | Female | 77 | 10 days | Open pit | |
| D4_TF_B | Female | 77 | 10 days | Open pit | |
| D4_TF_C | Female | 77 | 10 days | Open pit | |
| Post-deposition samples | |||||
| D1_TS_A | Female | 91 | 219 days | Open pit | |
| D1_TS_B | Female | 91 | 219 days | Open pit | |
| D1_TS_C | Female | 91 | 219 days | Open pit | |
| D2_TS_A | Female | 67 | 834 days | Burial | |
| D2_TS_B | Female | 67 | 834 days | Burial | |
| D2_TS_C | Female | 67 | 834 days | Burial | |
| D3_TS_A | Female | 61 | 790 days | Burial | |
| D3_TS_B | Female | 61 | 790 days | Burial | |
| D3_TS_C | Female | 61 | 790 days | Burial | |
| D4_TS_A | Female | 77 | 872 days | Open pit | |
| D4_TS_B | Female | 77 | 872 days | Open pit | |
| D4_TS_C | Female | 77 | 872 days | Open pit | |
Additional files
-
Supplementary file 1
Univariate analyses for all the individual omics.
- https://cdn.elifesciences.org/articles/83658/elife-83658-supp1-v2.xlsx
-
Supplementary file 2
Environmental data.
- https://cdn.elifesciences.org/articles/83658/elife-83658-supp2-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83658/elife-83658-mdarchecklist1-v2.docx
-
Source code 1
R pipeline employed in the study.
- https://cdn.elifesciences.org/articles/83658/elife-83658-code1-v2.zip