Osteosarcoma-enriched transcripts paradoxically generate osteosarcoma-suppressing extracellular proteins
Figures
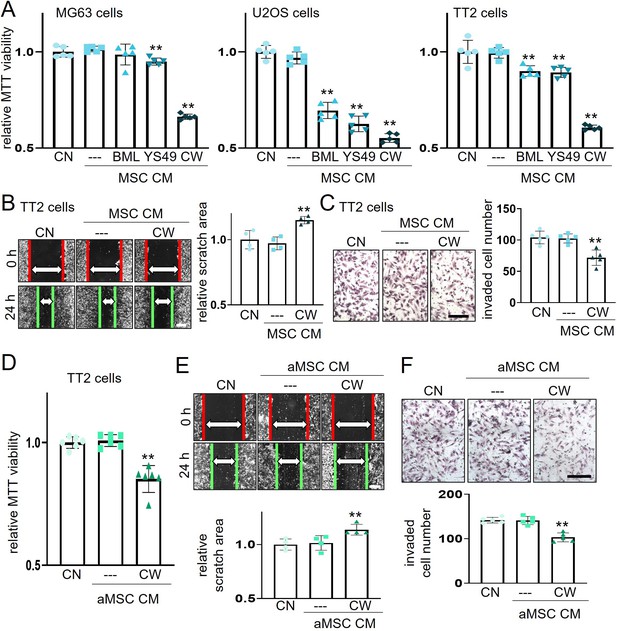
Suppression of the viability, migration, and invasion of osteosarcoma (OS) cells by CW008-treated mesenchymal stem cell (MSC) CM.
The double asterisk indicates p<0.01. CN = control, CM = conditioned medium, BML = BML284 as a Wnt activator, YS49=PI3K activator, CW = CW008 as a PKA activator, MSC = bone marrow-derived MSC, and aMSC = adipose-derived MSC. (A) Reduction in MTT-based cell viability of three OS cell lines (MG63, U2OS, TT2 PDX) in 2 days by bone marrow-derived MSC CM, which were derived after the treatment with BML284, YS49, or CW008. (n=5). (B and C) Decrease in scratch-based motility (n=4) and transwell invasion (n=5) of TT2 OS cells in 2 days by CW008-treated MSC CM. (D–F) Inhibition of MT-based cell viability (n=6), scratch-based motility (n=4), and transwell invasion (n=5) of TT2 OS cells in 2 days by CW008-treated adipose-derived MSC. (Scale bar, 200 µm, error bars indicate standard deviation.)
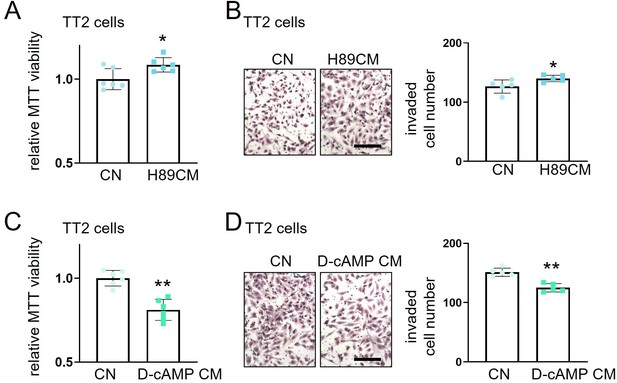
Effects of H89 (PKA inhibitor) and D-cAMP (cAMP analog as a PKA activator).
The single and double asterisks indicate p<0.05 and 0.01, respectively. CN = control, and CM = conditioned medium. (A and B) Elevation of MTT-based cell viability (n=6) and transwell invasion (n=5) of TT2 osteosarcoma (OS) cells by H89-treated MSC-derived CM. (C and D) Reduction of MTT-based cell viability (n=6) and transwell invasion (n=5) of TT2 OS cells by D-cAMP-treated MSC-derived CM. (Scale bar, 200 µm, error bars indicate standard deviation.)
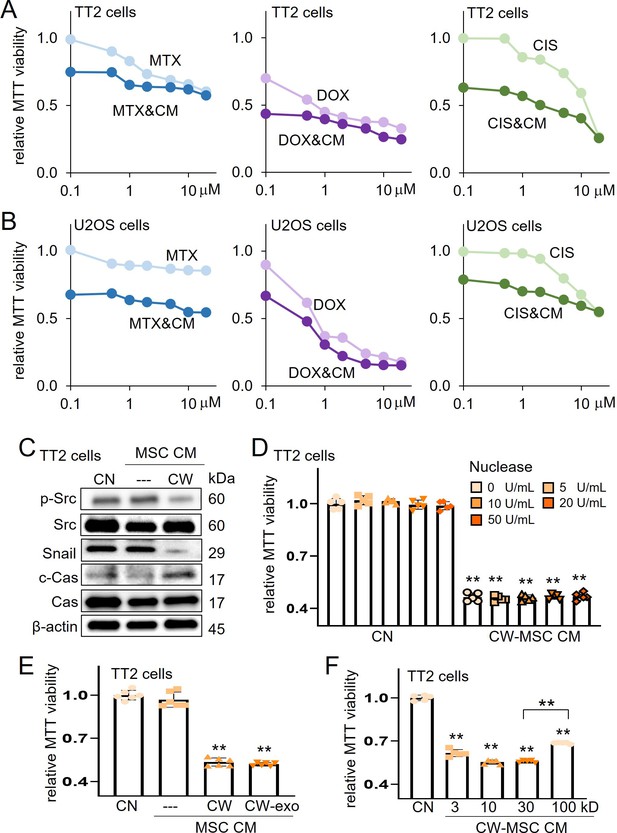
Characterization of CW008-treated mesenchymal stem cell (MSC) CM.
The double asterisk indicated p<0.01. CN = control, CW = CW008, CM = conditioned medium, Cas = caspase 3, exo = exosome, MTX = methotrexate, DOX = doxorubicin, and CIS = cisplatin. (A and B) Additive MTT-based anti-tumor effect of CW008-treated MSC CM with methotrexate, doxorubicin, and cisplatin in TT2 and U2OS cells, respectively. (C) Reduction of p-Src and Snail and elevation of cleaved caspase 3 in TT2 OS cells by CW008-treated MSC CM for 2 days. (D and E) No significant change of MTT-based viability by the nuclease treatment (n=5) and ultracentrifugation for exosome removal (n=6), respectively, of CW008-treated MSC CM. (F) Variable tumor-suppressing capability of the size-fractionated CW008-treated bone marrow-derived MSC CM (CW-MSC CM) portion. (n=5). The protein size in kD on the X-axis indicates the cutoff size. For instance, the bar for 100 kD indicates the MTT value for the fraction that includes proteins larger than 100 kD. (Error bars indicate standard deviation.)
-
Figure 2—source data 1
Original files for the gels in Figure 2C.
- https://cdn.elifesciences.org/articles/83768/elife-83768-fig2-data1-v1.zip
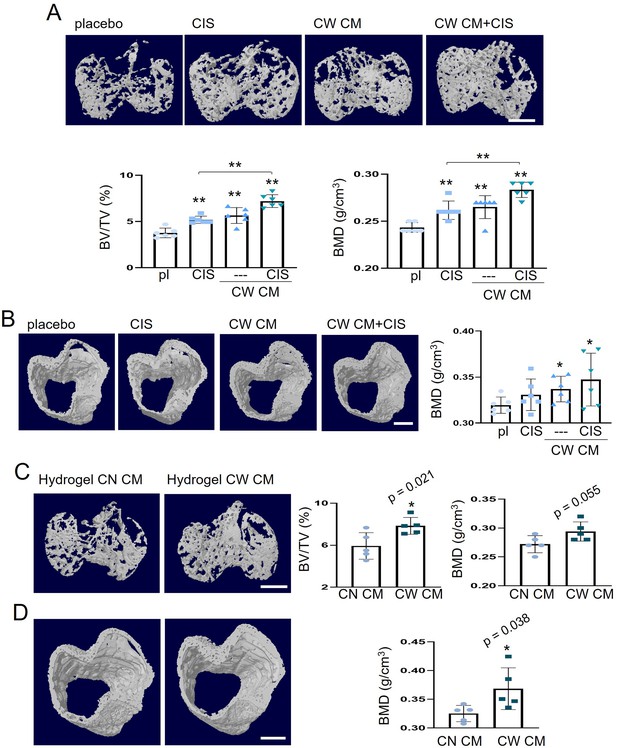
Protection of tumor-invaded bone by CW008-treated mesenchymal stem cell (MSC) CM.
pl = placebo, CIS = cisplatin, CW = CW008, CN = control, and CM = conditioned medium. The single and double asterisks indicate p<0.05 and 0.01, respectively. (A) Additive prevention of bone loss in the tumor-invaded tibia by CW008-treated MSC CM with cisplatin (n=6). (B) Effect of CW008-treated MSC CM with cisplatin on tibial cortical bone (n=6). (C and D) Reduction in trabecular and cortical bone loss in the tumor-invaded proximal tibia by hydrogel CW008-treated MSC CM. BV/TV = bone volume ratio, BMD = bone mineral density (n=5). (Scale bar, 1 mm, error bars indicate standard deviation).
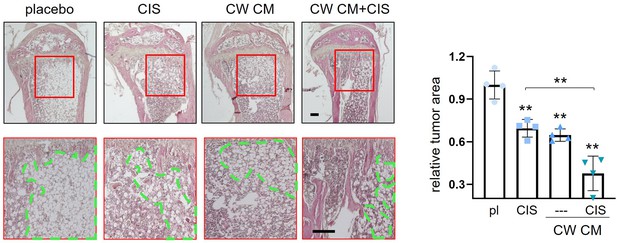
Histological analysis of tumor-invaded bone in response to CW008-treated MSC CM with and without cisplatin.
pl = placebo, CIS = cisplatin, CW = CW008, and CM = conditioned medium. The double asterisks indicate p<0.01. (n=4, scale bar, 200 µm, error bars indicate standard deviation.)
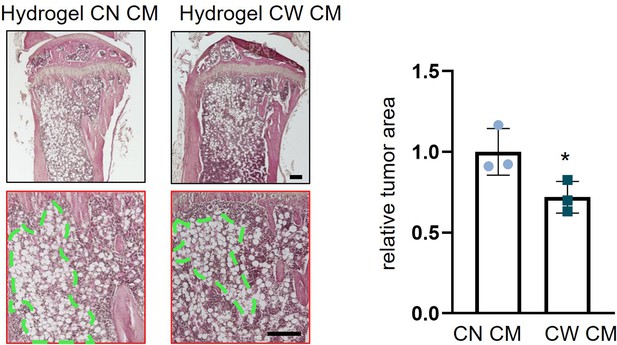
Histological analysis of reduction in trabecular bone loss in the tumor-invaded proximal tibia by hydrogel CW008-treated mesenchymal stem cell (MSC) CM.
CN = control, CW = CW008, and CM = conditioned medium. The single asterisk indicates p<0.05. (n=3, scale bar, 200 µm, error bars indicate standard deviation.)
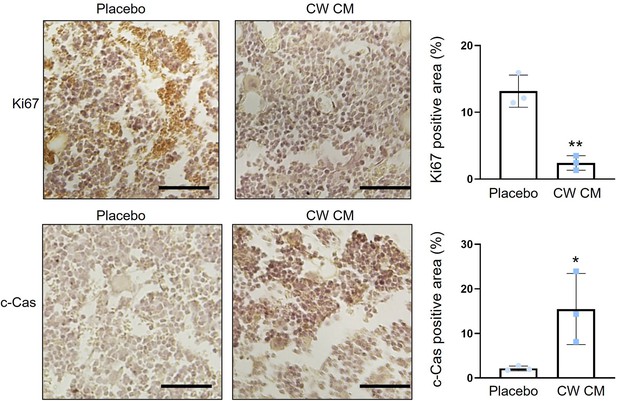
Downregulation of Ki-67 and upregulation of cleaved caspase 3 in tumor-invaded bone sections by the administration of CW CM.
CW CM = CW008-treated MSC CM, and c-Cas = cleaved caspase 3. The single and double asterisks indicate p<0.05 and 0.01, respectively. (n=3, scale bar, 50 µm, error bars indicate standard deviation.)
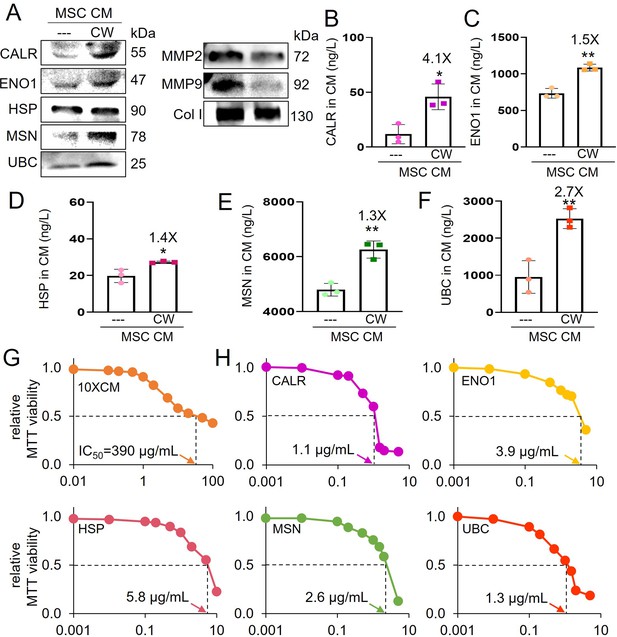
Tumor-suppressing proteins in CW008-treated mesenchymal stem cell (MSC) CM.
The single and double asterisks indicate p<0.05 and 0.01, respectively. CM = conditioned medium, CW = CW008 (PKA activator), CALR = calreticulin, ENO1=enolase 1, HSP = heat shock protein 90ab1, MSN = moesin, UBC = ubiquitin C, MMP2 and MMP9=matrix metalloproteinases 2 and 9, Col I=type I collagen. (A) Western blot-based expression levels of CALR, ENO1, HSP, MSN, UBC, MMP2, MMP9, and Col I in CW008-treated MSC CM. (B–F) ELISA-based levels of five tumor-suppressing proteins (CALR, ENO1, HSP, MSN, and UBC) in CW008-treated MSC CM (n=3). (G) Dose responses of TT2 OS cells in response to CW008-treated MSC CM with IC50 at 390 μg/ml (n=5). (H) Dose responses and IC50 of TT2 OS cells in response to five tumor-suppressing proteins (CALR, ENO1, HSP, MSN, and UBC). (Error bars indicate standard deviation.)
-
Figure 4—source data 1
Original files for the gels in Figure 4A.
- https://cdn.elifesciences.org/articles/83768/elife-83768-fig4-data1-v1.zip
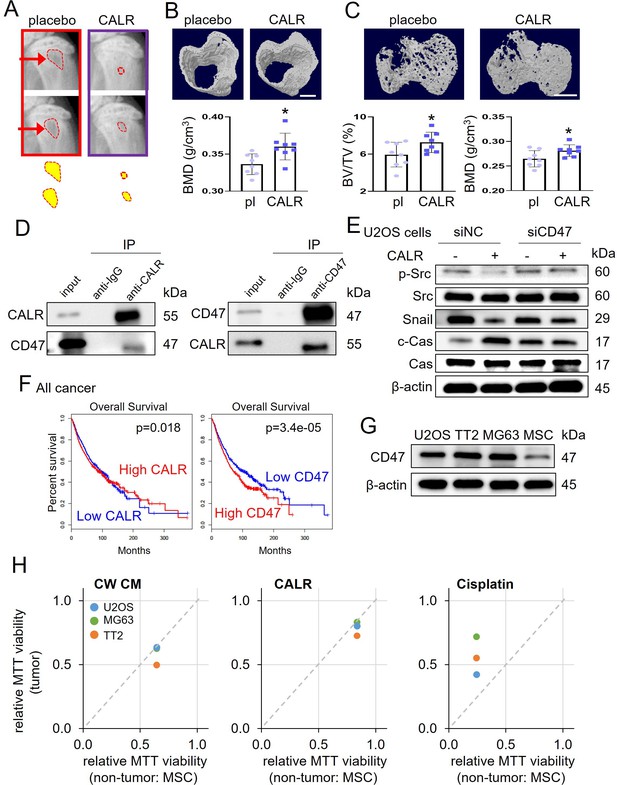
Calreticulin’s action on tumor-invaded bone and its interaction with CD47.
The single asterisk indicates p<0.05. CALR = calreticulin, pl = placebo, siNC = nonspecific siRNA, siCD47=CD47 siRNA, CW CM = CW008-treated MSC conditioned medium, and Cas = caspase 3. (A) X-ray images of the proximal tibia of NSG mice that received inoculation of TT2 OS cells. (B) MicroCT image-based increase in BMD (bone mineral density) of cortical bone in the proximal tibia (N=8) of NSG mice by daily injection of calreticulin (10 μg/kg). (C) MicroCT image-based increase in BV/TV (bone volume ratio) and BMD of trabecular bone of NSG mice in the proximal tibia (n=8). (D) Reciprocal co-immunoprecipitation of calreticulin and CD47 using TT2 osteosarcoma (OS) cell lysate. (E) Suppression of calreticulin-induced changes of p-Src, Snail, and cleaved caspase 3 (c-Cas) in U2OS cells by silencing CD47. (F) Favorable %survival with a high level of calreticulin and a low level of CD47 in all cancer patients. (G) Elevated expression level of CD47 in three OS cell lines (U2OS, TT2, and MG63), compared to non-OS cells (MSCs). (H) Tumor selectivity, larger than 1 for CW008-treated MSC CM and calreticulin, indicating the selective inhibition of OS cells (TT2, U2OS, and MG63) compared to MSCs. Of note, tumor selectivity for cisplatin, smaller than 1, indicates cisplatin’s non-selective inhibition of tumor and non-tumor cells. (Scale bar, 1 mm, error bars indicate standard deviation.)
-
Figure 5—source data 1
Original files for the gels in Figure 5D, E and G.
- https://cdn.elifesciences.org/articles/83768/elife-83768-fig5-data1-v1.zip
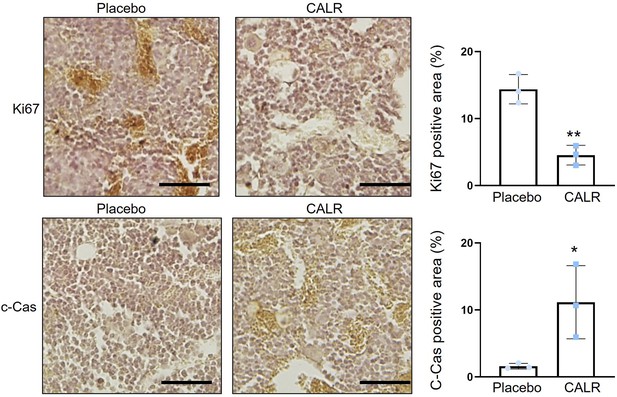
Downregulation of Ki-67 and upregulation of cleaved caspase 3 in tumor-invaded bone sections by the administration of calreticulin.
CALR = calreticulin, and c-Cas=cleaved caspase 3. The single and double asterisks indicate p<0.05 and 0.01, respectively. (n=3, scale bar, 50 µm, error bars indicate standard deviation.)
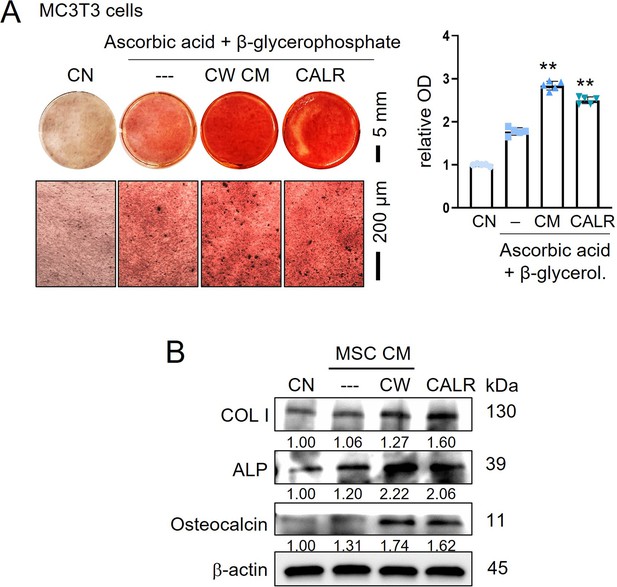
Stimulation of osteoblast differentiation by CW008-treated bone marrow-derived MSC CM (CW-MSC CM) and calreticulin.
CN = control, CW = CW008, CM = conditioned medium, CALR = calreticulin, COL I=type I collagen, and ALP = alkaline phosphatase. The double asterisks indicate p<0.01. (A) Enhanced Alizarin Red staining of MC3T3 osteoblasts by the administration of CW-MSC CM and calreticulin. (B) Elevation of type I collagen, ALP, and osteocalcin by CW-MSC CM and calreticulin. The double asterisks indicate p<0.01.
-
Figure 5—figure supplement 2—source data 1
Original files for the gels in Figure 5—figure supplement 2B.
- https://cdn.elifesciences.org/articles/83768/elife-83768-fig5-figsupp2-data1-v1.zip
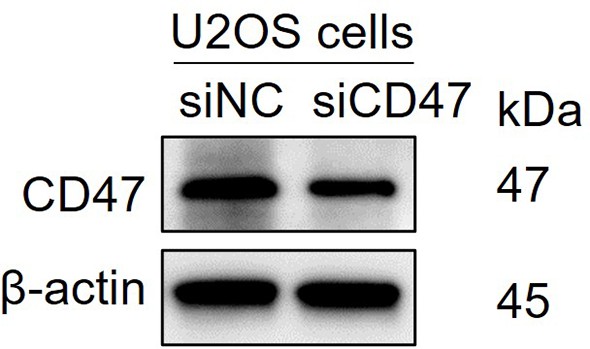
Silencing the CD47 in U2OS cells.
siNC = nonspecific control siRNA and siCD47=CD47 siRNA.
-
Figure 5—figure supplement 3—source data 1
Original files for the gels in Figure 5—figure supplement 3.
- https://cdn.elifesciences.org/articles/83768/elife-83768-fig5-figsupp3-data1-v1.zip
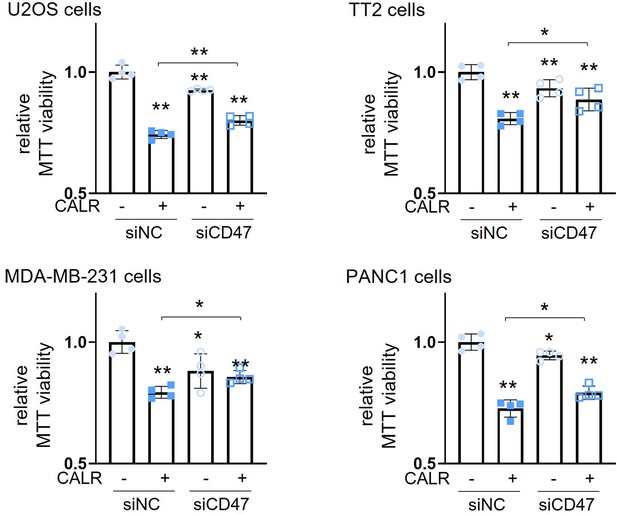
Suppression of CALR-driven decrease in MTT-based viability of U2OS osteosarcoma (OS) cells, TT2 OS cells, MDA-MB-231 breast cancer cells, and PANC1 pancreatic cells by partial silencing of CD47.
CALR = calreticulin, siNC = nonspecific control siRNA, and siCD47=CD47 siRNA. The single and double asterisks indicate p<0.05 and 0.01, respectively. (n=4, error bars indicate standard deviation.)
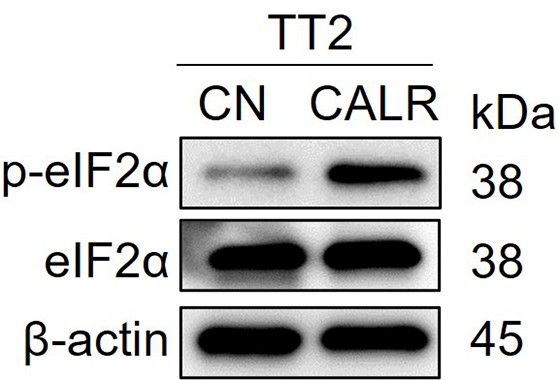
Elevation of the phosphorylation of eukaryotic translation initiation factor 2 alpha (p-eIF2a) in TT2 osteosarcoma (OS) cells by the application of calreticulin (CALR).
-
Figure 5—figure supplement 5—source data 1
Original files for the gels in Figure 5—figure supplement 5.
- https://cdn.elifesciences.org/articles/83768/elife-83768-fig5-figsupp5-data1-v1.zip
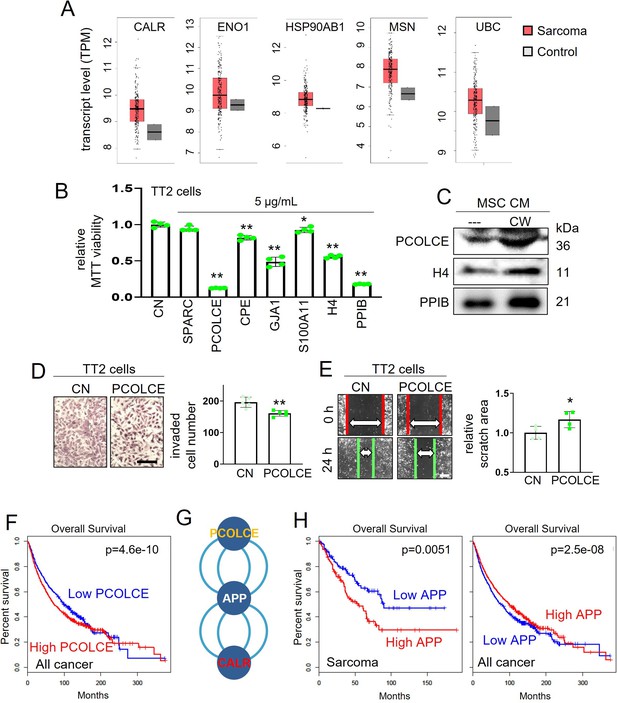
TCGA database-based prediction of tumor suppressors and the effect of PCOLCE.
CN = control and APP = amyloid precursor protein. The single and double asterisks indicate p<0.05 and 0.01, respectively. (A) List of five selected proteins that were expressed higher in sarcoma tissues than the normal tissues in the TCGA database. (B) Reduction in MTT-based proliferation of TT2 osteosarcoma (OS) cells by seven selected recombinant proteins (5 µg/ml) in 48 hr, which are selected with higher expression in sarcoma tissues in the TCGA database. (C) Western blot images, showing that the levels of H4, PPIB, and PCOLCE were elevated in CW008-treated bone marrow-derived MSC CM (CW-MSC CM). (D and E) Inhibition in the transwell invasion (48 hr) and scratch-based migration (24 hr) of TT2 OS cells by 1 µg/ml PCOLCE recombinant protein. (F) Lowered survival with a high level of PCOLCE in all cancer patients. (G) Protein candidates, which are connected with PCOLCE by IntAct network analysis. (H) Lowered survival rates of sarcoma patients with high levels of APP, while the opposite effect of all cancer types. (Scale bar, 200 µm, error bars indicate standard deviation.)
-
Figure 6—source data 1
Original files for the gels in Figure 6C.
- https://cdn.elifesciences.org/articles/83768/elife-83768-fig6-data1-v1.zip
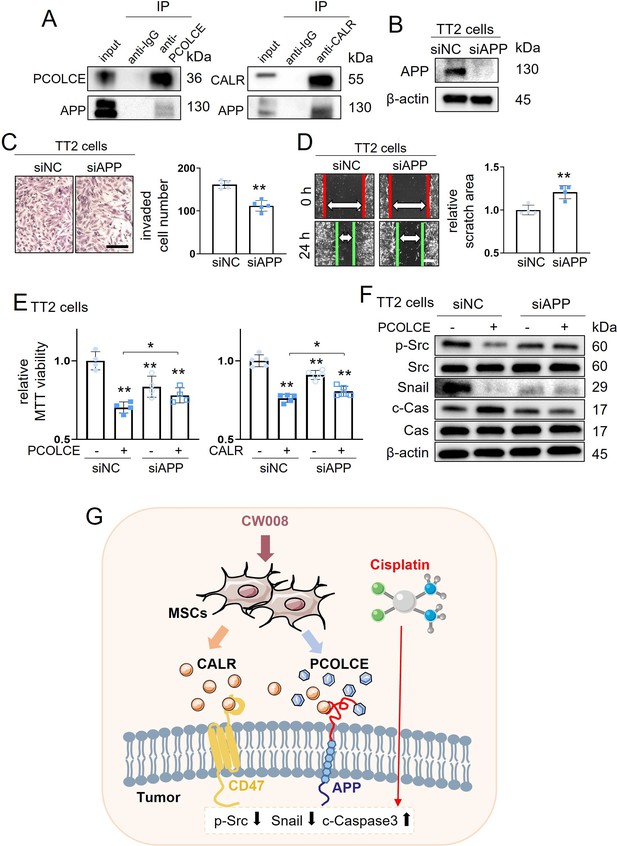
Putative regulatory mechanism for the tumor-suppressing capability of CW008-treated mesenchymal stem cell (MSC) conditioned medium (CM).
CALR = calreticulin, siNC = nonspecific siRNA, siAPP = APP siRNA, and Cas = caspase 3. The single and double asterisks indicate p<0.05 and 0.01, respectively. (A) Co-immunoprecipitation of amyloid precursor protein (APP) with PCOLCE and CALR. (B–D) Inhibition in the transwell invasion (48 hr) (n=5) and scratch-based migration (24 hr) (n=4) of TT2 osteosarcoma (OS) cells by silencing the APP. (E) Suppression of PCOLCE (n=4) and CALR (n=5)-driven decrease in MTT-based viability of TT2 OS cells by partial deletion of APP. (F) Suppression of PCOLCE-induced changes of p-Src and cleaved caspase 3 (c-Cas) in TT2 cells by silencing APP. (G) Schematic diagram for the regulatory mechanism of tumor-suppressing action of CW008-treated MSC CM. (Scale bar, 200 µm, error bars indicate standard deviation.)
-
Figure 7—source data 1
Original files for the gels in Figure 7A, B and F.
- https://cdn.elifesciences.org/articles/83768/elife-83768-fig7-data1-v1.zip
Additional files
-
Supplementary file 1
List of seven selected proteins that were expressed higher in sarcoma tissues than the normal tissues in the TCGA dataset.
- https://cdn.elifesciences.org/articles/83768/elife-83768-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83768/elife-83768-mdarchecklist1-v1.docx





