MeCP2 regulates Gdf11, a dosage-sensitive gene critical for neurological function
Figures
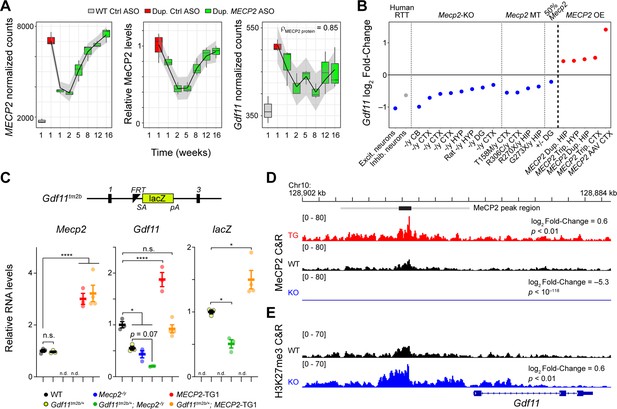
Gdf11 is positively regulated by MeCP2.
(A) Dynamic expression levels of MECP2, MeCP2, and Gdf11 during anti-MECP2 antisense oligonucleotide (ASO) treatment in MECP2 duplication mice (data from Shao et al., 2021b). Spearman correlation coefficient between Gdf11 and MeCP2 is shown in the right panel. Gray interval represents a loess fit ± standard error. (B) Log2 fold-change of GDF11 expression in excitatory and inhibitory neurons isolated from post mortem brains from individuals with Rett syndrome and Gdf11 expression in Mecp2-knockout (Mecp2-KO), mutant (Mecp2 MT), and MECP2 overexpression (MECP2 OE) mouse model RNA-seq experiments. Abbreviations used to describe studies are as follows: CTX: cortex, CB: cerebellum, HYP: hypothalamus, DG: dentate gyrus, HIP: hippocampus, Dup.: duplication, Trip.: triplication, AAV: adeno-associated virus. Grayed color indicates padjusted >0.1. Studies are ordered from 1 to 20 and correspond to the study number in Supplementary file 1. (C) A Gdf11-knockout first allele (Gdf11tm2b) has a LacZ cassette with splice acceptor and polyA sequence knocked in between exons 1 and 3 of Gdf11, and exon 2 of Gdf11 is deleted. Quantitative PCR of Mecp2, Gdf11, and the LacZ reporter from the cerebellum of wild-type, Gdf11tm2b/+, Mecp2-/y, Gdf11tm2b/+; Mecp2-/y, MECP2-TG1, and Gdf11tm2b/+; MECP2-TG1 (n=3–4 biological replicates per genotype). n.d. denotes the given gene expression measurement was not detected. qPCR data is shown as mean ± sem. (D) CUT&RUN profiling of MeCP2 binding in MECP2 transgenic mice (MECP2-TG1), wild-type, and Mecp2-knockout hippocampi (Mecp2-KO) at the Gdf11 locus. Black bar represents a MeCP2 peak called by MACS2 software, and the gray bar represents an expanded region of ±3.5 kb of increased MeCP2 binding. Log2 fold-change of MeCP2 occupancy within the gray region is shown relative to wild-type, and the p-value of the comparison is shown beneath the fold-change. Tracks are displayed as an aggregate of biological replicates (n=3–6 biological replicates per genotype). (E) CUT&RUN profiling of H3K27me3 in wild-type and Mecp2-knockout (Mecp2-KO) at the Gdf11 locus. Log2 fold-change of binding within the gray region is shown relative to wild-type, and the p-value of the comparison is shown beneath the fold-change. Tracks are displayed as an aggregate of biological replicates (n=3 biological replicates per genotype). (*) p<0.05, (****) p<0.0001 from one way ANOVA followed by multiple comparisons testing.
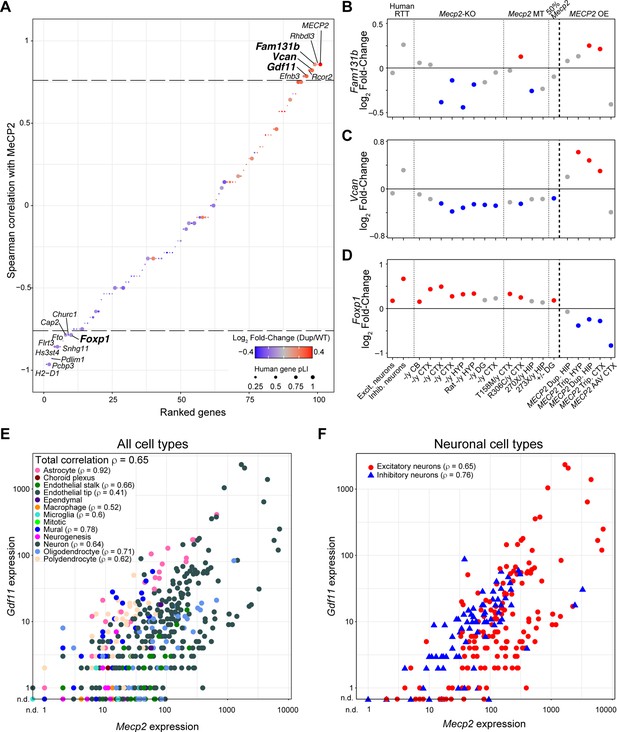
Gene expression changes correlated with MeCP2 protein levels.
(A) Spearman correlation of genes misregulated in MECP2 duplication mice quantifying how they track with MeCP2 levels during anti-MECP2 antisense oligonucleotide treatment (Shao et al., 2021b). Data points are colored by log2 fold-change between MECP2 duplication and wild-type mice. The size of the dot represents the probability of loss intolerance (pLI) of the human gene if an exact match exists from the gnomad database. Genes with correlation above a threshold of 0.75 (horizontal line) are labeled by name. Genes with pLI >0.9 are bolded. (B–C) Fam131b, Vcan, and Foxp1 are MeCP2-correlated genes predicted to be intolerant to loss-of-function but are not consistently misregulated in excitatory and inhibitory neurons isolated from post mortem brains from individuals with Rett syndrome and expression in Mecp2-knockout (Mecp2-KO), mutant (Mecp2 MT), and MECP2 overexpression (MECP2 OE) mouse model RNA-seq experiments (see Figure 1B). Abbreviations used to describe studies are as follows: CTX: cortex, CB: cerebellum, HYP: hypothalamus, DG: dentate gyrus, HIP: hippocampus, Dup.: duplication, Trip.: triplication, AAV: adeno-associated virus. Dots denote log2 fold-change between MeCP2 perturbed tissue and wild type, and gray color indicates padj >0.1. Studies are ordered from 1 to 20 and correspond to the study number in Supplementary file 1. (E–F) Cell-type specific correlation of Gdf11 and Mecp2. Single-cell expression of Gdf11 and Mecp2 was extracted from the DropViz database and split into the annotated cell types. (E) The total Spearman’s correlation across all cell types is shown in the upper left and the Spearman’s correlation for each cell type is denoted next to the legend label for each cell type. Correlations were not calculated for cell-types with fewer than 20 observations. (F) The neuron subtype was split into excitatory and inhibitory neurons using Slc17a7 expression (see Materials and methods). Spearman’s correlation for each neuronal type is denoted next to the legend label for each neuron type.
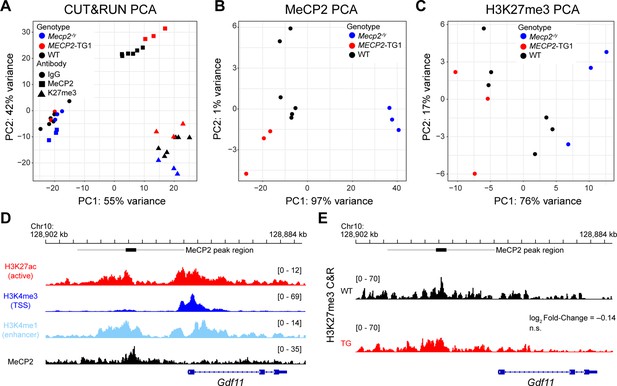
Epigenetic changes correlated with MeCP2 protein levels.
(A–C) Global assessment of MeCP2 and H3K27me3 binding in Mecp2-knockout, wild-type, and MECP2-TG1 hippocampi assessed by principal component analysis (PCA). (A) PCA of MeCP2 (square), H3K27me3 (triangle), and IgG (circle) CUT&RUN profiles in Mecp2-knockout (blue), wild-type (black), and MECP2-TG1 (red) hippocampi. (B) PCA of MeCP2 binding profiles only colored by genotype. (C) PCA of H3K27me3 binding profiles only colored by genotype. (D) Histone modifications, as measured by ChIP-seq, denoting open chromatin (H3K27ac), the transcriptional start site (H3K4me3), and putative enhancers (H3K4me1) from the hippocampus are shown alongside the MeCP2 binding profile around the Gdf11 locus. (E) CUT&RUN profiling of H3K27me3 in wild-type and MECP2-TG1 at the Gdf11 locus. Black bar represents a MeCP2 peak called by MACS2 software, and the gray bar represents an expanded region of ±3.5 kb. Log2 fold-change of binding within the gray region is shown relative to wild-type, and the p-value of the comparison is shown beneath the fold-change (n=3 biological replicates per genotype).
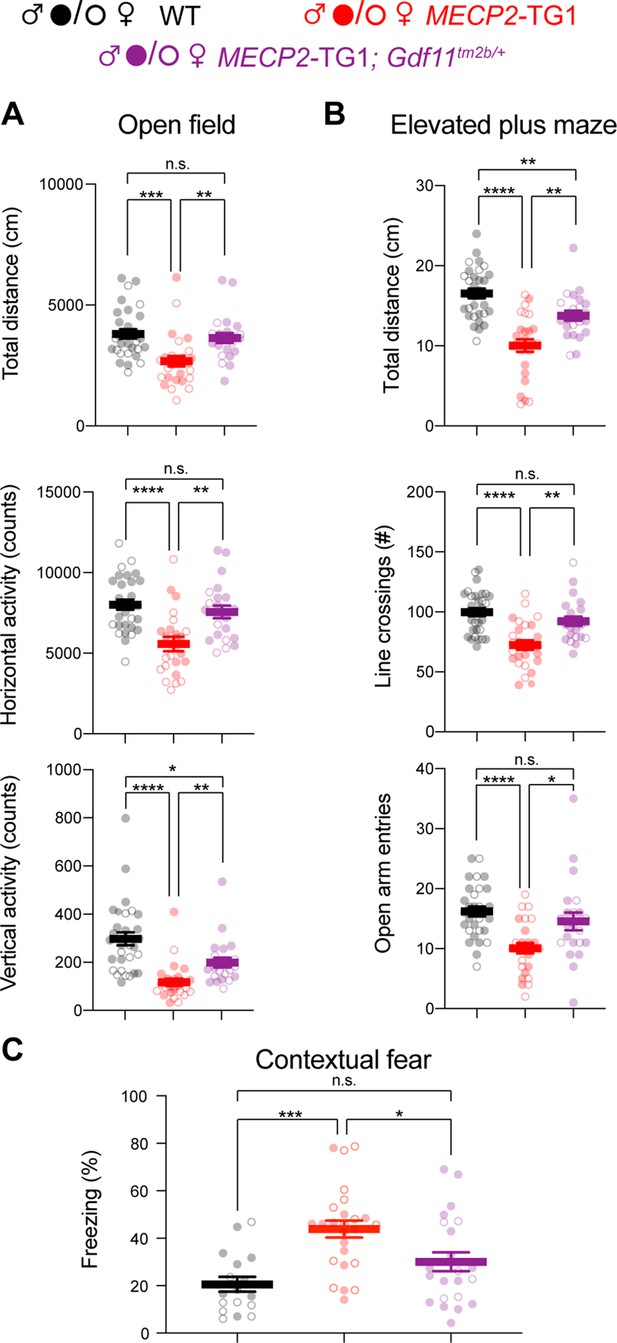
Genetic reduction and normalization of Gdf11 dose ameliorates several behavioral deficits in MECP2-TG1 mice.
Behavioral characterization of MECP2-TG1, MECP2-TG1; Gdf11tm2b/+ double mutants, and their respective wild-type littermate controls was performed beginning at 16 weeks of age. (A) Open-field assessment of locomotion and activity. (B) Elevated plus maze assay measures of movement and anxiety-like behaviors. (C) Learning assessment using contextual fear-conditioning. Greater freezing indicates better memory of the context. Central estimate of data is shown as mean ± sem. Closed circles denote male mouse data points and open circles denote female mouse data points. For all open field and elevated plus maze (A,B), n=30 wild-type mice (19 male, 11 female); n=26 MECP2-TG1 mice (14 male, 12 female); and n=22 MECP2-TG1; Gdf11tm2b/+ mice (16 male, 6 female). For fear conditioning assay (C), n=17 wild-type mice (9 male, 8 female); n=25 MECP2-TG1 mice (13 male, 12 female); and n=22 MECP2-TG1; Gdf11tm2b/+ mice (16 male, 6 female). All data were analyzed using a Welsch one-way ANOVA with Dunnett’s post hoc multiple comparisons, and raw measurements are provided in Figure 2—source data 1. * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001.
-
Figure 2—source data 1
Raw data from all behavioral assays related to Figure 2, Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/83806/elife-83806-fig2-data1-v1.xlsx
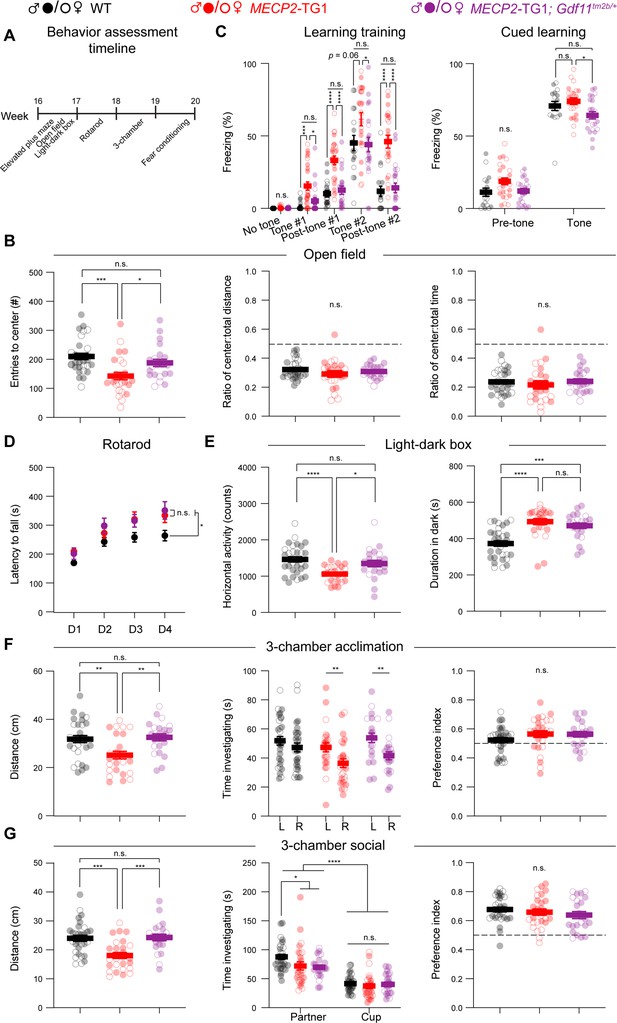
Correction of Gdf11 dose does not ameliorate all behavioral deficits in MECP2-TG1 mice.
Behavioral characterization of MECP2-TG1, MECP2-TG1; Gdf11tm2b/+ double mutants, and their respective wild-type littermate controls was performed beginning at 16 weeks of age. (A) Timeline and order of behavioral assays. (B) Open-field assessment of anxiety-like behavior as measured by entries into the center of the arena, ratio of the distance traveled in the center to total, and ratio of the time spent in the center over total. (C) Learning assessment during training and cued fear-conditioning. Greater freezing indicates better memory of the cue. (D) Motor coordination measured by the rotating rod assay. (E) Locomotion and anxiety-like behaviors measured by the light-dark assay. Mice with anxiety-like behaviors prefer to spend longer duration in the dark segment of the arena. (F, G) Sociability measured by the three-chamber assay, split into acclimation phase (F) and social phase (G). Central estimate of data is shown as mean ± sem. Closed circles denote male mouse data points and open circles denote female mouse data points. For all assays except fear conditioning, n=30 wild-type mice (19 male, 11 female); n=26 MECP2-TG1 mice (14 male, 12 female); and n=22 MECP2-TG1; Gdf11tm2b/+ mice (16 male, 6 female). For fear conditioning assay (C), n=17 wild-type mice (9 male, 8 female); n=25 MECP2-TG1 mice (13 male, 12 female); and n=22 MECP2-TG1; Gdf11tm2b/+ mice (16 male, 6 female). All data were analyzed using a Welsch one way ANOVA with Dunnett’s post hoc multiple comparisons, and raw measurements are provided in Figure 2—source data 1. * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001.
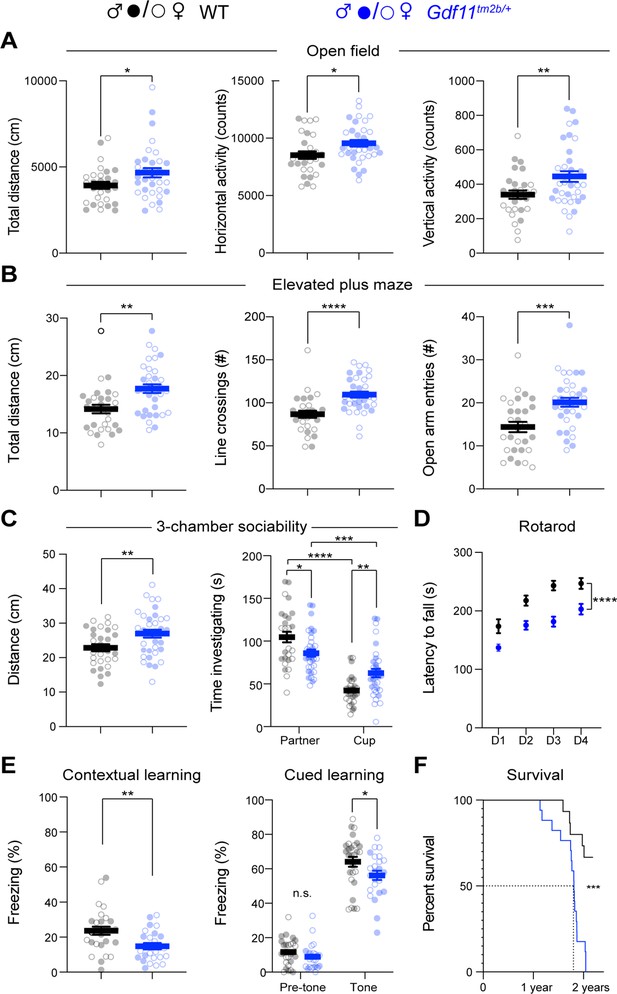
Mice lacking one copy of Gdf11 (Gdf11tm2b/+) display behavioral deficits.
Behavioral characterization of Gdf11tm2b/+ mice and their respective wild-type littermate controls was performed beginning at 16 weeks of age. (A) Open field assessment of locomotion and activity. (B) Elevated plus maze assay measures of movement and anxiety-like behaviors. (C) Distance traveled and time investigating novel mouse in 3-chamber sociability assay. (D) Motor coordination measured by the rotating rod assay. (E) Learning assessment using contextual and cued fear-conditioning. Greater freezing indicates better memory of the context. (F) Survival analysis. Central estimate of data is shown as mean ± sem. Closed circles denote male mouse data points and open circles denote female mouse data points. For behavioral measurements except fear conditioning, n=29 wild-type mice (15 male, 14 female) and n=34 Gdf11tm2b/+ mice (16 male, 18 female). For fear conditioning assay, n=28 wild-type mice (16 male, 12 female) and n=24 Gdf11tm2b/+ mice (9 male, 15 female). For survival analysis, n=15 wild-type mice and n=17 Gdf11tm2b/+ mice. Behavioral data were analyzed using Welsch t-test and raw measurements are provided in Figure 3—source data 1. Survival was analyzed using Mantel-Cox test. * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001.
-
Figure 3—source data 1
Raw data from all behavioral assays related to Figure 3 and Figure 3—figure supplement 2.
- https://cdn.elifesciences.org/articles/83806/elife-83806-fig3-data1-v1.xlsx
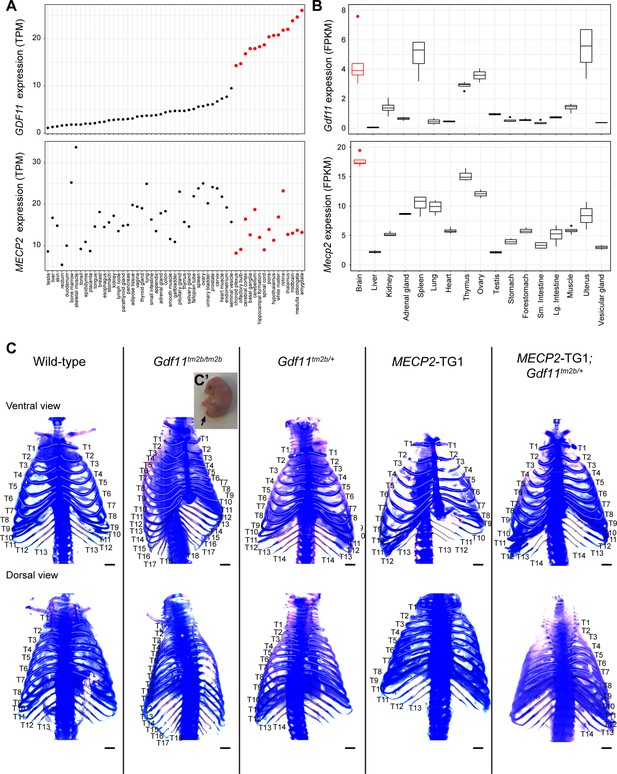
Gdf11 is highly expressed in the brain and loss of one copy causes abnormal skeletal development.
(A) GDF11 expression is higher in the brain compared to peripheral tissues. The consensus human tissue expression dataset from the Human Protein Atlas was queried for GDF11 (top) and MECP2 (bottom) expression levels by tissue. Tissues are sorted based on GDF11 expression and tissues in the central nervous system are colored red. (B) Gdf11 is highly expressed in the mouse brain. Mouse tissue RNA-seq expression dataset was taken from Li et al., 2017 and queried for Gdf11 (top) and Mecp2 (bottom) expression levels by tissue. Brain samples are colored red. Data from n=4 replicates is shown as a boxplot. (C) Gdf11tm2b/tm2b and Gdf11tm2b/+ mice exhibit the same skeletal patterning defects as described in the original Gdf11-/- line. Widefield imaging of the rib segments from P0 pups of the indicated genotype using Alcian blue and Alizarin red staining. Attached ribs are highlighted with white dotted lines, while unattached ribs are highlighted with black dotted lines. The top panels show the skeleton from a ventral view, while the bottom panels show the skeleton from a dorsal view. Image is representative of n=13 (WT), 4 (Gdf11tm2b/tm2b), 24 (Gdf11tm2b/+), 4 (MECP2-TG1), and 4 (MECP2-TG1; Gdf11tm2b/+) pups. Scale bar is 2 mm. (C’, inset) Image of P0 Gdf11tm2b/tm2b pup displaying truncated tail and perinatal lethality.
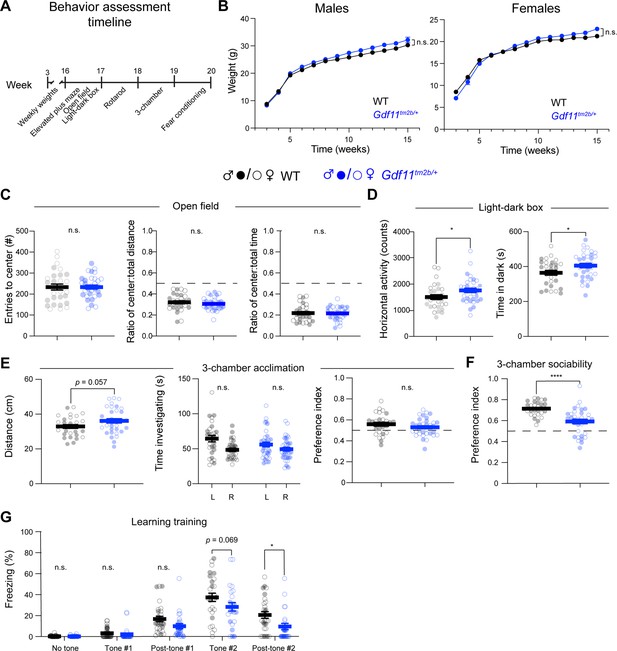
Loss of one copy of Gdf11 does not alter all behavioral phenotypes.
Behavioral characterization of Gdf11tm2b/+ mice and their respective wild-type littermate controls was performed beginning at 16 weeks of age. (A) Timeline and order of behavioral assays. (B) Weight measurements in male and female Gdf11tm2b/+ and wild-type littermates. Left plot displays data for males, and right plot displays data for females. (C) Open field assessment of anxiety-like behavior as measured by entries into the center of area and ratios of distance and time spent in the center. (D) Locomotion and anxiety-like behavior measured by the light-dark assay. Mice with anxiety-like behaviors prefer to spend longer duration in the dark segment of the arena. (E,F) Sociability measured by the 3-chamber assay, split into acclimation phase (E) and social phase presented as the ratio of time investigating partner mouse over total time investigating (F). (G) Learning during shock-tone fear conditioning paradigm. Central estimate of behavior data is shown as mean ± sem. Closed circles denote male mouse data points and open circles denote female mouse data points. For behavioral measurements except fear conditioning, n=29 wild-type mice (15 male, 14 female) and n=34 Gdf11tm2b/+ mice (16 male, 18 female). For fear conditioning assay, n=28 wild-type mice (16 male, 12 female) and n=24 Gdf11tm2b/+ mice (9 male, 15 female). For survival analysis, n=15 wild-type mice and n=17 Gdf11tm2b/+ mice. Behavioral data were analyzed using Welsch t-test, and raw measurements are provided in Figure 3—source data 1. * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001.
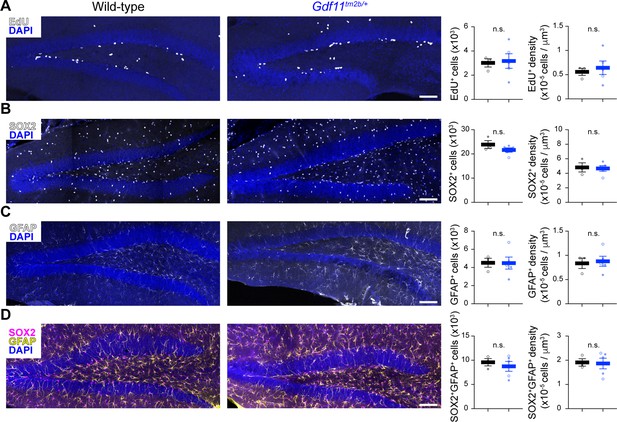
Loss of one copy of Gdf11 does not alter adult neurogenesis in the dentate gyrus.
Quantification of markers of (A) proliferative cells (EdU) or (B–D) neural progenitor pools (SOX2 and GFAP) in the subgranular zone of the dentate gyrus in wild-type or Gdf11tm2b/+ mice. Representative images of the dentate gyrus for the indicate stains are shown on the left. The projected total number of cells and the density of cells is shown on the plots to the right using stereology to quantify n=3–6 separate slices per animal (N=3 wild-type animals and 5 Gdf11tm2b/+ animals) (see Methods). Each data point is the aggregated value for one animal. Closed circles denote male animals and open circles denote female animals. Data are presented as mean ± sem. Data were analyzed using Welsch’s t-test, and raw measurements are provided in Figure 4—source data 1. Scale bar is 200 μm and is the same for all images.
-
Figure 4—source data 1
Raw measurements and stereology quantification of neurogenesis markers.
- https://cdn.elifesciences.org/articles/83806/elife-83806-fig4-data1-v1.xlsx
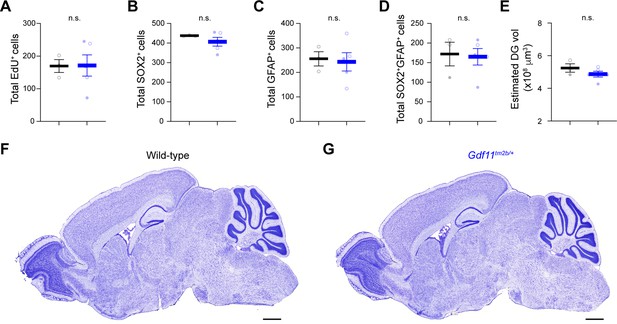
Loss of one copy of Gdf11 does not cause gross anatomical abnormalities or gross differences in progenitor pools.
(A–D) Total number of EdU (A), SOX2 (B), GFAP (C), or SOX2; GFAP double positive (D) cells scored by stereology. Counts are sums from n=3–6 separate slices per animal (N=3 wild-type animals and 5 Gdf11tm2b/+ animals). (E) Estimated volume of the dentate gyrus from stereological quantification. (F,G) Images of Cresyl violet stain of sagittal sections from wild-type or Gdf11tm2b/+ brains. Scale bar is 1 mm. Each data point is the sum for one animal. Closed circles denote male animals and open circles denote female animals. Data are presented as mean ± sem. Data were analyzed using Welsch’s t-test, and raw measurements are provided in Figure 4—source data 1.

Partner side does not change preference index during social phase in Gdf11tm2b/+ mice.
Data from Figure 3 - figure supplement 2F were split into conditions with partner mice on left versus right and trends are preserved in this representation too. Data were analyzed with two-way ANOVA and post-hoc multiple comparisons. (****) p < 0.0001 between genotypes.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus; male and female) | Gdf11tm2a(EUCOMM)Hmgu | MMRRC | Cat. #037721-UCD; RRID: MMRRC_037721-UCD | Gdf11tm2b allele generated by breeding with Sox2-Cre; maintained on C57BL/6J background |
| Strain, strain background (Mus musculus; male and female) | MECP2-TG1 (Tg(MECP2)1Hzo) | The Jackson Laboratory | Cat. #008679; RRID: IMSR_JAX:008679 | Maintained on C57BL/6J background |
| Strain, strain background (Mus musculus; male and female) | Mecp2tm1.1Bird | The Jackson Laboratory | Cat. #003890; RRID: IMSR_JAX:003890 | Maintained on C57BL/6J background |
| Strain, strain background (Mus musculus; female) | Sox2-Cre | The Jackson Laboratory | Cat. #008454; RRID: IMSR_JAX:008454 | |
| Strain, strain background (Mus musculus; male and female) | C57BL/6J | The Jackson Laboratory | Cat. #000664; RRID: IMSR_JAX:000664 | |
| Antibody | Rabbit anti-MeCP2 (clone D4F3) (rabbit monoclonal) | Cell Signaling Technology | Cat. #3456; RRID: AB_2143894 | 1:100 |
| Antibody | Rabbit anti-Try-Methyl-Histone H3 (Lys27) (clone C36B11) (rabbit monoclonal) | Cell Signaling Technology | Cat. #9733; RRID: AB_2616029 | 1:100 |
| Antibody | Rabbit IgG (rabbit polyclonal) | Millipore | Cat. #12–370; RRID: AB_145841 | 1:100 |
| Antibody | Mouse anti-GFAP (mouse monoclonal) | Sigma-Aldrich | Cat. #G3893; RRID: AB_477010 | 1:1000 |
| Antibody | Rabbit anti-SOX2 (rabbit polyclonal) | Abcam | Cat. #ab97959; RRID: AB_2341193 | 1:500 |
| Antibody | Donkey anti-rabbit Alexa 488 (donkey polyclonal) | Jackson ImmunoResearch | Cat #711-545-152; RRID: AB_2313584 | 1:1000 |
| Antibody | Donkey anti-mouse Alexa 555 (donkey polyclonal) | ThermoFisher Scientific | Cat #A-31570; RRID: AB_2536180 | 1:1000 |
| Other | Concanavalin A magnetic beads | Bangs Laboratories Inc | Cat. #BP531 | See Materials and Methods, CUT&RUN |
| Other | pAG-MNase | Epicypher | Cat. #15–1016 | See Materials and Methods, CUT&RUN |
| Other | RNAse A | ThermoFisher Scientific | Cat. #EN0531 | See Materials and Methods, CUT&RUN |
| Other | E. coli spike-in | Epicypher | Cat. #18–1401 | See Materials and Methods, CUT&RUN |
| Other | MaXtract tubes | Qiagen | Cat. #129046 | See Materials and Methods, CUT&RUN |
| Commercial assay or kit | NEB Next II DNA Ultra Kit | New England Biolabs | Cat. #E7645S | |
| Commercial assay or kit | Unique Combinatorial Dual Index kit | New England Biolabs | Cat. #E6442S | |
| Other | SPRI select beads | Beckman Coulter | Cat. #B23318 | See Materials and Methods, CUT&RUN Next Generation Sequencing Library Preparation |
| Commercial assay or kit | KAPA library quantification kit | Roche | Cat. #5067 | |
| Other | 5-ethynyl-2’-deoxyuridine (EdU) | Invitrogen | Cat. #E10187 | See Materials and Methods, EdU and Immunofluorescnece staining |
| Commercial assay or kit | Click-iT EdU Imaging Kit with AlexaFluor 647 dye | Invitrogen | Cat. #C10340 | |
| Other | Alcian blue | Sigma-Aldrich | Cat. #A5268 | See Materials and Methods, Skeletal analysis |
| Other | Alizarin red | Sigma-Aldrich | Cat. #A5533 | See Materials and Methods, Skeletal analysis |
| Software, algorithm | GraphPad Prism 9 | GraphPad Software | https://www.graphpad.com | |
| Software, algorithm | Trimmomatic v 0.36 | GitHub | https://github.com/usadellab/Trimmomatic; RRID: SCR_011848 | Version 0.36 |
| Software, algorithm | Bowtie2-2.3.4.1 | GitHub | https://github.com/BenLangmead/bowtie2; RRID: SCR_016368 | Version 2.3.4.1 |
| Software, algorithm | Bedtools | GitHub | https://github.com/arq5x/bedtools2; RRID: SCR_00646 | Version v2.29.1 |
| Software, algorithm | deeptools | GitHub | https://github.com/deeptools/deepTools; RRID: SCR_016366 | Version 3.3.0 |
| Software, algorithm | MACSr | GitHub | https://github.com/macs3-project/MACSr; RRID: SCR_013291 | MACS peak calling using R wrapper; Version 1.0.0 |
Additional files
-
Supplementary file 1
Summary and source of transcriptomics studies mined in this study.
- https://cdn.elifesciences.org/articles/83806/elife-83806-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83806/elife-83806-mdarchecklist1-v1.docx





