Inhibition of the proton-activated chloride channel PAC by PIP2
Figures
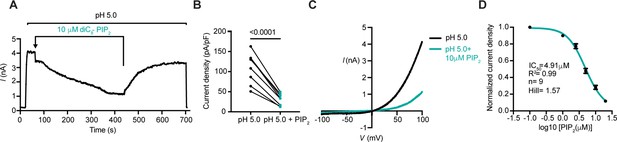
PIP2 inhibits the PAC channel activity.
(A) Representative whole-cell current trace at + 100 mV (2 s/sweep) showing inhibition of endogenous PAC currents by bath perfusion of soluble diC8-PIP2 at 10 μM concentration. (B) PAC current densities before and after application of PIP2 for 150 s. Statistical significance was determined using a two-tailed Student’s paired t-test. (C) Representative I/V curve of pH 5-induced PAC currents before and after PIP2 treatment. (D) Dose-dependent inhibition of pH 5-induced PAC currents by PIP2 yielded a half-maximal inhibition, IC50, of 4.91 μM with a Hill slope of 1.57. Bars are reported as mean ± SEM.
-
Figure 1—source data 1
Data and statistics plotted in Figure 1.
- https://cdn.elifesciences.org/articles/83935/elife-83935-fig1-data1-v2.xlsx
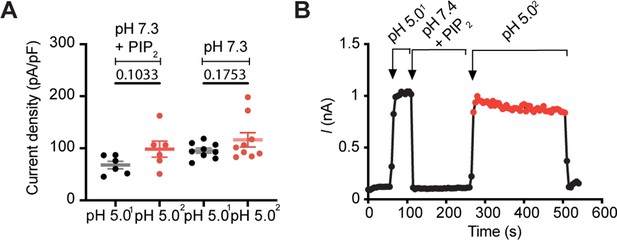
PIP2 does not bind to the closed PAC channel.
(A) Current amplitude was measured at pH 5.0 before (1) and after (2) perfusion of diC8-PIP2 at pH 7.3 (+PIP2). The control cells were perfused with pH 7.3 only. There was no significant difference in current density before and after treatment of cells with PIP2 at neutral pH. Statistical significance was determined using a two-tailed Student’s unpaired t-test. (B) Representative current trace at + 100 mV (3 s/sweep) of the experiment described in (A).
-
Figure 1—figure supplement 1—source data 1
Data and statistics plotted in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/83935/elife-83935-fig1-figsupp1-data1-v2.xlsx
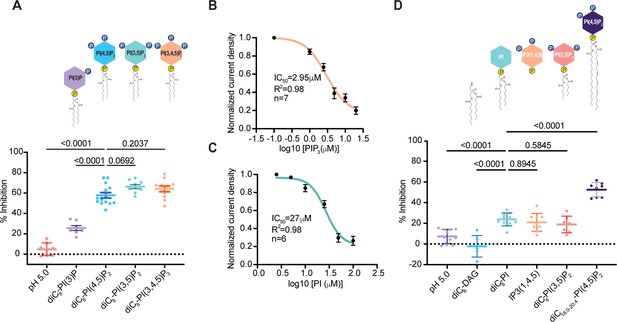
Phosphates and acyl chain length synergistically contribute to PAC inhibition by PIP2.
(A) Percent inhibition of pH 5-induced PAC currents by different diC8-phosphatidylinositol lipids at 10 μM concentration: PI(3)P, PI(4,5)P2, PI(3,5)P2, and PI(3,4,5)P3. Statistical significance was determined using ordinary one-way ANOVA with the Dunnett post hoc test. Bars are reported as mean ± SEM. (B) Dose-dependent inhibition of PAC currents by PIP3. Bars are reported as mean ± SEM. (C) Dose-dependent inhibition of PAC currents by phosphatidylinositol (PI). Bars are reported as mean ± SEM. (D) Percent inhibition of pH 5-induced PAC currents by phosphatidylinositol lipids of different acyl chain length 10 μM concentration: Diacylglycerol (DAG), diC8-PI, IP3(1,4,5), diC6-PI(3,5)P2 and diC18:0-20:4-PI(4,5)P2. Statistical significance was assessed using ordinary one-way ANOVA with the Dunnett post hoc test. Bars are reported as mean ± SEM.
-
Figure 2—source data 1
Data and statistics plotted in Figure 2.
- https://cdn.elifesciences.org/articles/83935/elife-83935-fig2-data1-v2.xlsx
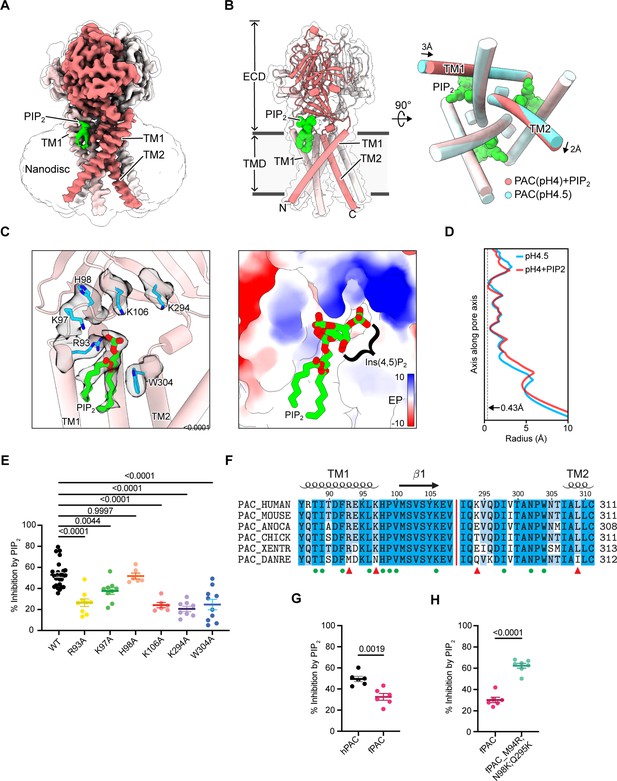
PIP2 binds directly to the PAC channel.
(A) Cryo-EM structure of PAC channel at pH 4.0 with bound PIP2. One subunit is shown in red with TM1 and TM2 labeled. The density corresponding to putative PIP2 is colored green. (B) The structural model of PAC channel at pH 4.0 with bound PIP2 in side view (left) and bottom-up view (right). For comparison, the PAC channel at pH 4.5 without PIP2 (PDBID: 7SQH) is shown in cyan for the bottom-up view (right). (C) A close-up view of the PIP2 binding site. (Left) Cartoon representation of the PIP2 binding site. Important residues relevant to the study of the putative PIP2 binding site, including R93, K97, H98, K106, K294, and W304, are shown in stick. Cryo-EM densities for PIP2 and the nearby residues are shown in a semi-transparent surface. The Ins(4,5)P2 group is not resolved in the Cryo-EM map and thus not modeled in the deposited structure. (Right) Surface representation of the PIP2 binding site colored with electrostatic potential. Unit in kcal/mol/e-. A full PIP2 molecule, including the hypothetically positioned Ins(4,5)P2 group, is shown in the right panel. (D) The pore profile of PAC at pH 4.0 with PIP2 (PDBID: 8FBL) and at pH 4.5 without PIP2 (PDBID: 7SQH). The smallest radius along the pore axis is 0.43 Å, suggesting that both structures are impermeable to chloride ions. (E) Mutating PIP2-binding residues to alanine significantly decreases diC8-PIP2-mediated inhibition on pH 5-induced PAC currents. The constructs were expressed in PAC KO HEK293 cells for recordings. Statistical significance was assessed using one-way ANOVA with the Dunnett post hoc test. Bars are reported as mean ± SEM. (F) Multiple sequence alignment of several PAC orthologs. Key residues that form the PIP2 binding site are labeled using green dots. Binding site residues that are not conserved in zebrafish PAC (PAC_DANRE) are indicated by red triangles. (G) Percent inhibition (mean ± SEM) of hPAC or fPAC current at pH 5.0 by 10 μM diC8-PIP2. Zebrafish PAC (fPAC) shows significantly less inhibition by PIP2 compared to human PAC. Statistical significance was determined using a two-tailed Student’s unpaired t-test. (H) Mutating zebrafish PAC residues to the corresponding human PAC residues, M94R, N98K and Q295K, significantly increases the inhibition by PIP2 in comparison to the wild-type zebrafish PAC. Statistical significance was determined using a two-tailed Student’s unpaired t-test. Bars are reported as mean ± SEM.
-
Figure 3—source data 1
Data and statistics plotted in Figure 3.
- https://cdn.elifesciences.org/articles/83935/elife-83935-fig3-data1-v2.xlsx
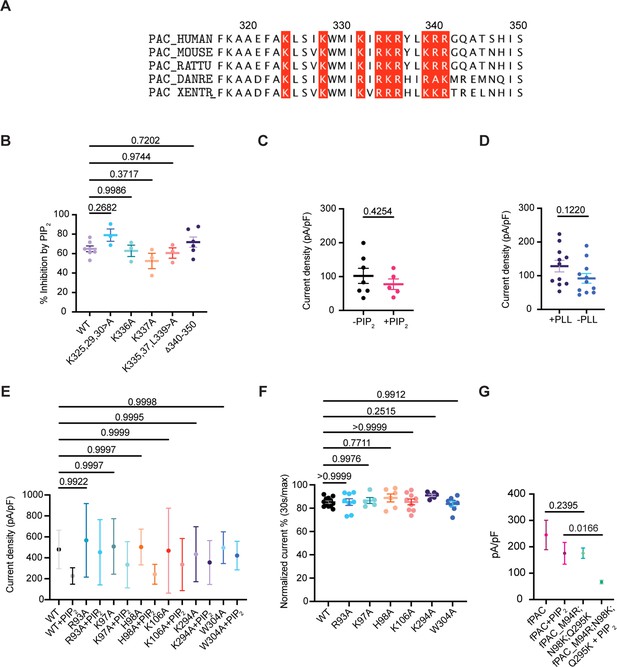
The binding site for PIP2 is not on the intracellular side of the PAC channel.
(A) We hypothesized that a cluster of positively charged residues at the C-terminus of PAC, outlined in red, binds PIP2. (B) Potential cytosolic PIP2-binding residues were screened by making grouped alanine mutations, or single alanine mutants, and by deleting a 10 amino-acid sequence at the C-terminal end of PAC. There was no significant difference in PIP2 inhibition on pH 5.0-induced PAC currents when the mutants were overexpressed in PAC knockout HEK293 cells. Statistical significance was determined using one-way ANOVA with Dunnett post hoc test. Bars are reported as mean ± SEM. (C) 10 μM diC8-PIP2 added through a patch pipette containing intracellular solution (ICS) in whole-cell configuration showed no significant difference when compared to ICS without PIP2 in HEK293 cells. Statistical significance was determined using a two-tailed Student’s unpaired t-test. Bars are reported as mean ± SEM. (D) Current density before and after depleting endogenous PIP2 from the inner leaflet by applying 100 μg/ml of Poly-L-Lysine (PLL) through the patch pipette in HEK293 cells. Statistical significance was determined using a two-tailed Student’s unpaired t-test. Bars are reported as mean ± SEM. (E) Current density before adding PIP2 is unaffected when PIP2-binding residues were mutated to alanine in hPAC. Statistical significance was determined using one-way ANOVA with the Dunnett post hoc test. Bars are reported as mean ± SEM. (F) Minimal desensitization (mean ± SEM) of PIP2-binding mutants at pH 5.0. Desensitized current after 30 s of acidic exposure was normalized to the initial maximum current of that recording and expressed as a percentage. Statistical significance was determined using one-way ANOVA with the Dunnett post hoc test. (G) Current density is unchanged when fPAC residues are mutated to the corresponding hPAC residues, while it is significantly decreased in the presence of PIP2. Statistical significance was determined using a two-tailed Student’s unpaired t-test. Bars are reported as mean ± SEM.
-
Figure 3—figure supplement 1—source data 1
Data and statistics plotted in Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/83935/elife-83935-fig3-figsupp1-data1-v2.xlsx
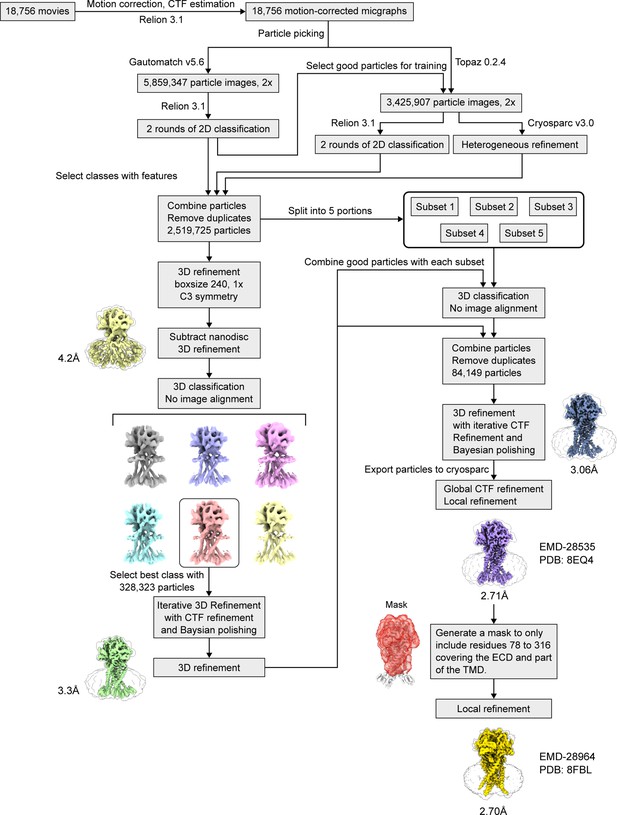
The cryo-EM data processing workflow of human PAC in nanodisc at pH 4.0 with 0.5 mM PIP2 dataset.
A more detailed description of this process can be found in the Materials and methods section. The refined map displayed in the workflow contains a transparent outline that is made of the unsharpened PAC map refined without using a mask. This is solely for visualization purposes such that the nanodisc signal (indicative of the transmembrane domain) can be seen more obviously.
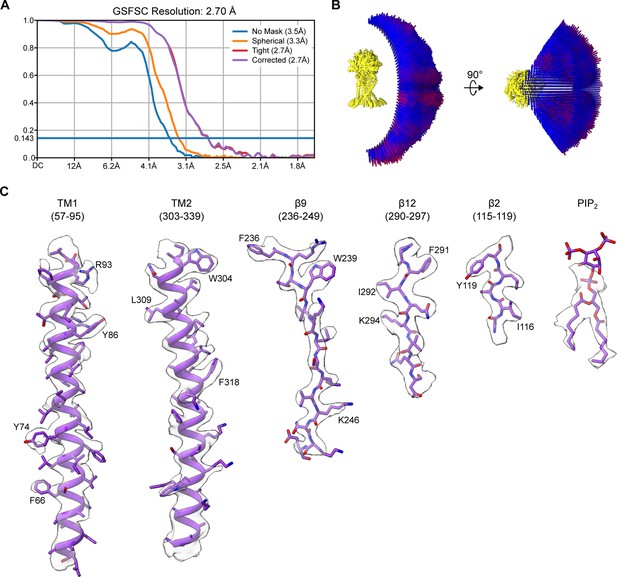
The reconstruction metrics of human PAC in nanodisc at pH 4.0 with 0.5 mM PIP2.
(A) The gold-standard Fourier shell correlation curve of the final cryo-EM map (EMD-28964). (B) The angular distribution of particles that give rise to the final reconstruction. (C) The representative densities of the reconstruction map, including TM1/2, β2, β9, β12, and PIP2.
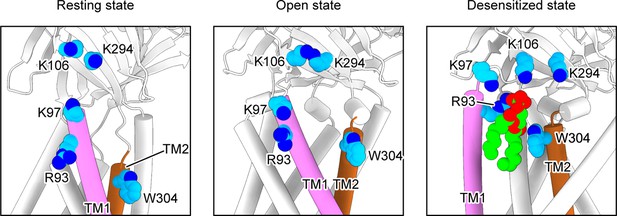
Putative PIP2-binding residues mapped in the PAC structures in the resting, open, and desensitized states, respectively.
The putative PIP2 binding site is made by the adjacent TM1 and TM2, which are colored in magenta and brown, respectively. Relevant residues for PIP2 binding are colored in blue.
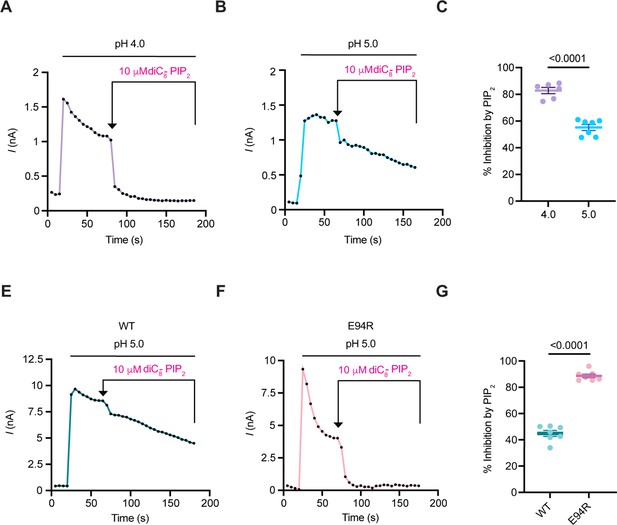
PIP2-mediated PAC inhibition correlates with the degree of channel desensitization.
(A, B) Representative current traces at + 100 mV (5 s/sweep) of endogenous PAC currents at pH 4.0 and 5.0 treated with 10 μM diC8-PIP2. diC8-PIP2 was applied after desensitized current reached a plateau. (C) Percent inhibition (mean ± SEM) of PAC currents at pH 4.0 and 5.0, 100 s after perfusion of 10 μM diC8-PIP2. Statistical significance was determined using a two-tailed Student’s unpaired t-test. (D, E) Representative current traces at + 100 mV (5 s/sweep) of overexpressing PAC WT and E94R at pH 5.0 treated with 10 μM diC8-PIP2. (F) Percent inhibition (mean ± SEM) of PAC WT and E94R currents at pH 5.0, 100 s after perfusion of 10 μM diC8-PIP2. Statistical significance was determined using a two-tailed Student’s unpaired t-test.
-
Figure 4—source data 1
Data and statistics plotted in Figure 4.
- https://cdn.elifesciences.org/articles/83935/elife-83935-fig4-data1-v2.xlsx
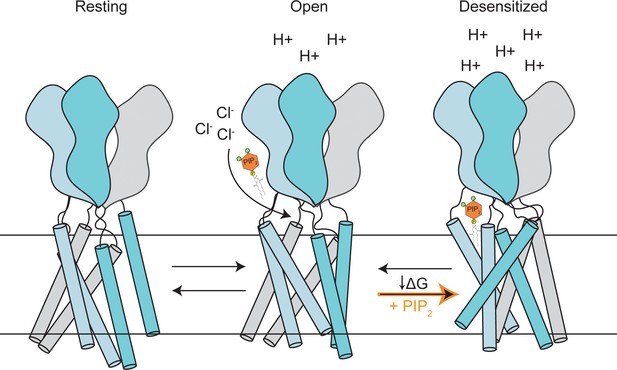
A proposed model of PAC inhibition by PIP2.
PAC channel adopts resting/open/desensitized states depending on the acidity of the environment. PIP2 selectively binds and stabilizes the desensitized conformation of PAC on the extracellular side of the membrane, altering the conformational/free energy landscape of the channel. As a result, in the presence of PIP2, a significant portion of PAC will be restricted in the desensitized conformation, leading to channel inhibition.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | hPAC | doi:10.1126/science.aav9739 | NP_060722/Q9 H813 | |
| Gene (Danio rerio) | fPAC | doi:10.1126/science.aav9739 | NP_001278691/Q7SY31 | |
| Recombinant DNA reagent | pEGC-hPAC (plasmid) | doi:10.1038/s41586-020-2875-7 | ||
| Recombinant DNA reagent | pIRES2-EGFP-hPAC (plasmid) | doi:10.1126/science.aav9739 | ||
| Recombinant DNA reagent | pIRES2-EGFP-fPAC (plasmid) | This paper | In the cell culture section of Materials and methods in this paper | |
| Recombinant DNA reagent | pEGC-hPAC | doi:10.1038/s41586-020-2875-7 | ||
| Cell line (Homo sapiens) | HEK293T | ATCC | Cat#:CRL-3216 | |
| Cell line (Homo-sapiens) | tsA-201 | Sigma Aldrich | Cat#: 85120602 | Cell line (Homo-sapiens) |
| Cell line (Homo sapiens) | PACC1 KO HEK293T | doi:10.1126/science.aav9739 | ||
| Chemical compound, drug | 08:0 PI (1,2-dioctanoyl-sn-glycero-3-phospho-(1'-myo-inositol) (ammonium salt)) | Avanti Polar Lipids | Cat#:850181 P | |
| Chemical compound, drug | 08:0 PI(4,5)P2 (1,2-dioctanoyl-sn-glycero-3-phospho-(1'-myo-inositol-4',5'-bisphosphate) (ammonium salt)) | Avanti Polar Lipids | Cat#:850185 P | |
| Chemical compound, drug | 08:0 PI(3,5)P2 (1,2-dioctanoyl-sn-glycero-3-phospho-(1'-myo-inositol-3',5'-bisphosphate) (ammonium salt)) | Avanti Polar Lipids | Cat#:850184 P | |
| Chemical compound, drug | 08:0 PI(3)P (1,2-dioctanoyl-sn-glycero-3-(phosphoinositol-3-phosphate) (ammonium salt)) | Avanti Polar Lipids | Cat#:850187 P | |
| Chemical compound, drug | 06:0 PI(3,5)P2 (1,2-dihexanoyl-sn-glycero-3-phospho-(1'-myo-inositol-3',5'-bisphosphate) (ammonium salt)) | Avanti Polar Lipids | Cat#:850174 P | |
| Chemical compound, drug | 18:0-20:0- PI(4,5)P2 (1-stearoyl-2-arachidonoyl-sn-glycero-3-phospho-(1'-myo-inositol-4',5'-bisphosphate)) (ammonium salt) | Avanti Polar Lipids | Cat#:850165 P | |
| Chemical compound, drug | IP3(1,4,5) (D-myo-inositol-1,4,5-triphosphate (ammonium salt)) | Avanti Polar Lipids | Cat#:850115 P | |
| Chemical compound, drug | 08:0 DG (1,2-dioctanoyl-sn-glycerol) | Avanti Polar Lipids | Cat#:800800O | |
| Chemical compound, drug | Poly-L-Lysine (PLL) | Sigma-Aldrich | Cat#:26124-78-7 | |
| Commercial assay or kit | Lipofectamine 2000 | Invitrogen | Cat#:11668–019 | |
| Commercial assay or kit | QuikChange II XL site-directed mutagenesis | Agilent Technologies | Cat#:200522 | |
| Software, algorithm | Clampfit 10.7 | Molecular devices | ||
| Software, algorithm | GraphPad Prism 9 | GraphPad | ||
| Software, algorithm | Clustal Omega | https://www.ebi.ac.uk/Tools/msa/clustalo/ | ||
| Software, algorithm | Relion | doi:10.7554/eLife.42166 | ||
| Software, algorithm | Cryosparc | doi:10.1038/nmeth.4169 | ||
| Software, algorithm | MotionCor2 | doi:10.1038/nmeth.4193 | ||
| Software, algorithm | ChimeraX | doi:10.1002/pro.3943 | ||
| Software, algorithm | CTFFIND4 | doi:10.1016/j.jsb.2015.08.008 |
Additional files
-
Supplementary file 1
Cryo-EM data collection, refinement, and validation statistics.
- https://cdn.elifesciences.org/articles/83935/elife-83935-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83935/elife-83935-mdarchecklist1-v2.docx





