High-throughput proteomics of nanogram-scale samples with Zeno SWATH MS
Figures
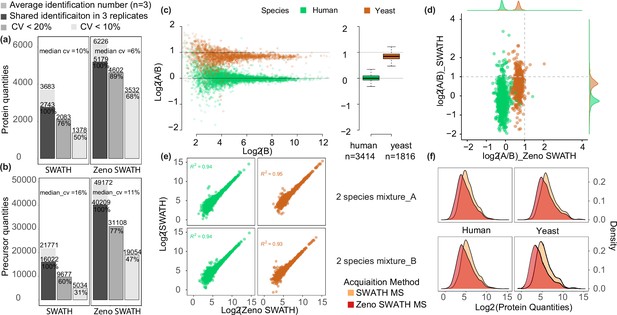
Comparison between SWATH and Zeno SWATH MS in proteome coverage and quantification precision and using 5 µl/min, 20 min micro-flow-rate chromatography.
(a,b) Reproducibility of protein identification using SWATH MS and Zeno SWATH MS on human cell line standard (K562) separated by micro-flow chromatography with a 62.5 ng load. Average identification numbers of proteins (a) and precursors (b) across three technical replicates (grey background bar) from a human cell line (K562) proteome tryptic digest are given; numbers of consistent identifications in technical replicates are given in dark grey; proteins or precursors quantified with coefficient of variation (CV) better than 20% in grey, and those quantified with a CV better than 10% in light grey. (c) Protein-level LFQbench results for Zeno SWATH MS. Quantification precision was benchmarked using yeast proteome tryptic digests that were spiked in two different proportions (A and B, three repeat injections each) into human cell line standard (K562) (A: 30 ng K562 + 35 ng yeast; B: 30 ng K562 + 17.5 ng yeast). Raw data were processed by library-free mode DIA-NN analysis. Protein ratios between the mixtures were visualised using the LFQbench R package (Navarro et al., 2016). Black dashed lines represent the expected log2(A/B) values for human (=0) and yeast (=1). Left pane, log-transformed ratios (log2(A/B)) of proteins plotted for DIA-NN over the log-transformed intensity of sample B. Right pane, protein quantification performance shown as box plots (boxes, interquartile range; whiskers, 1–99 percentile; n=3414 [human] and n=1816 [yeast]). (d) Average protein quantity differences between mixture A and B. Compared to SWATH MS, Zeno SWATH MS shows an improved separation of the two-species proteomes and more accurately represents the real ratio of the precursors (log2 of human quantity ratio close to 0, while log2 of yeast quantity ratio close to 1). (e) Correlation of protein quantities between both MS schemes in different mixtures. Human and yeast protein quantities in both mixtures show a high correlation between methods. (f) Distribution of protein quantities of each species in each mixture (A and B). In both mixtures and both species, Zeno SWATH MS shows deeper proteomic coverage by quantifying more low-abundant proteins.
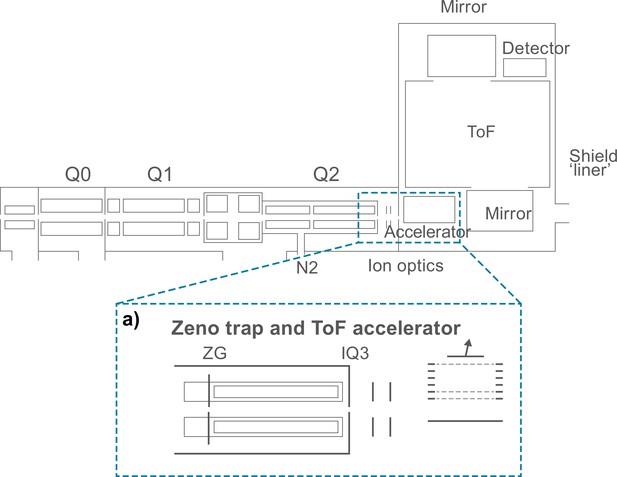
The ZenoTOF structure and Zeno Trap used in Zeno SWATH.
(a) Expanded view of the Zeno trap – a linear ion trap introduced after the collision cell (Q2). When enabled, all fragment ions in the selected mass range are trapped in an axial pseudopotential well created by an additional RF (‘AC’) voltage applied with the same amplitude and phase to all four rods of the trap in a required focal point. The release of ions from this trap is by potential energy with timing aligned to the next pulse at the accelerator.
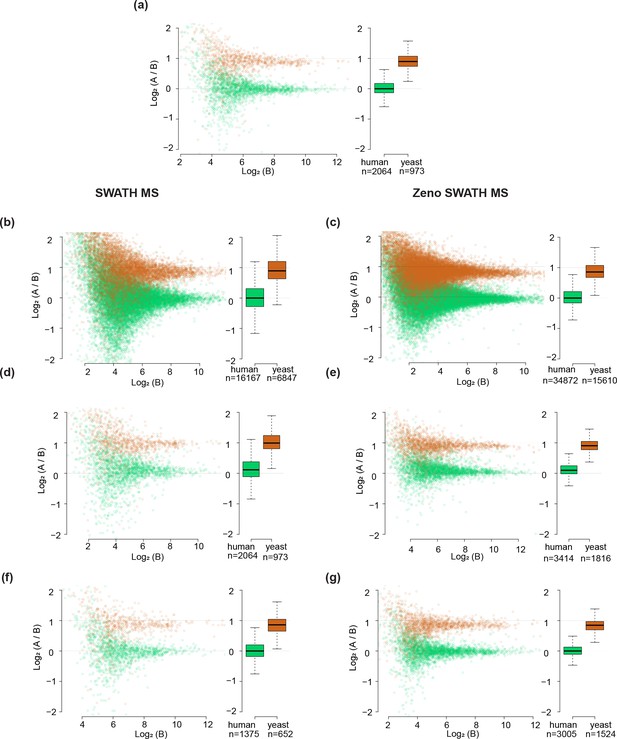
Comparison between SWATH MS and Zeno SWATH MS in quantification precision with two-species proteome mixture analysis.
(a) Protein-level LFQbench results for SWATH MS. (b) Precursor-level LFQbench results for SWATH MS. (c) Precursor-level LFQbench results for Zeno SWATH MS upon the separation of 1 µl injection of mixtures (A: 30 ng K562 + 35 ng yeast; B: 30 ng K562 + 17.5 ng yeast). (d) Protein-level LFQbench results for SWATH MS without normalisation (PG.Quantity was used). (e) Protein-level LFQbench results for Zeno SWATH MS without normalisation (PG.Quantity was used). (f) Protein-level LFQbench with a 50% lower amount (0.5 µl injection of mixtures [A: 30 ng K562 + 35 ng yeast; B: 30 ng K562 + 17.5 ng yeast]) results for SWATH MS. (g) Protein-level LFQbench with a 50% lower amount results for Zeno SWATH MS.
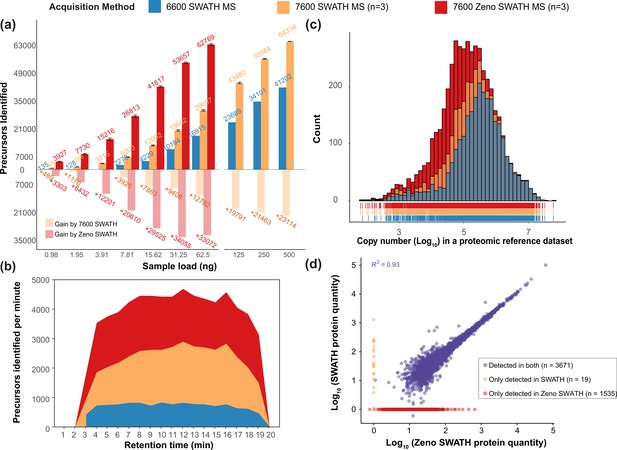
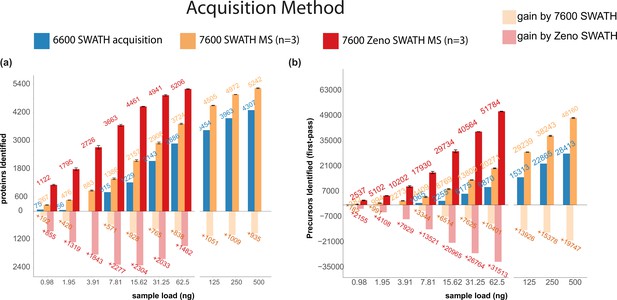
Zeno SWATH MS and its performance on K562 human cell line standard using 5 µl/min, 20 min micro-flow chromatography (SWATH MS on the TripleTOF 6600 system [blue], SWATH MS on the ZenoTOF 7600 system [yellow], and Zeno SWATH MS on the ZenoTOF 7600 system [red]).
(a) Precursor identification performance using SWATH and Zeno SWATH. Illustrated is the average number of precursors identified (n=3, error bars [±1 SD] shown in plot) for a K562 standard dilution series under three acquisition methods with library-free DIA-NN analysis (second-pass data). (b) Visualisation of precursor identification across gradient time with SWATH and Zeno SWATH MS of 62.5 ng K562 standard injection. (c) Histogram (bins = 50) of the protein abundance distribution represented as copy number in log10 scale of a human proteomic reference dataset (Bekker-Jensen et al., 2017). Zeno SWATH MS increases protein identification numbers by quantifying more low-abundant proteins. (d) Correlation of protein quantity between SWATH and Zeno SWATH MS. Illustrated is the correlation of protein quantity between two MS acquisition schemes; 3671 proteins. proteins were identified and quantified in both acquisition schemes, 19 proteins only quantified with SWATH MS, 1535 proteins quantified only by Zeno SWATH MS. Among the 3671 proteins quantified by both acquisition schemes, there is a high correlation of protein quantities between the proteins identified in both acquisition schemes (r2=0.93.).
Zeno SWATH MS and its performance on the K562 human cell line standard using 5 µl/min, 20 min micro-flow chromatography (SWATH MS on the TripleTOF 6600 system [blue], SWATH MS on the ZenoTOF 7600 system [yellow], and Zeno SWATH MS [red]).
(a) Protein identification performance using SWATH and Zeno SWATH MS. Illustrated is the average number of proteins identified (n=3, error bars [±1 SD]) for a K562 human cell line standard dilution series under three acquisition methods with library-free DIA-NN analysis (second-pass data). (b) Precursor identification performance using SWATH and Zeno SWATH MS. Illustrated is the average number of precursors identified (n=3, error bars [±1 SD] shown in plot) for a K562 human cell line standard dilution series under three acquisition methods with library-free DIA-NN analysis (first-pass data).
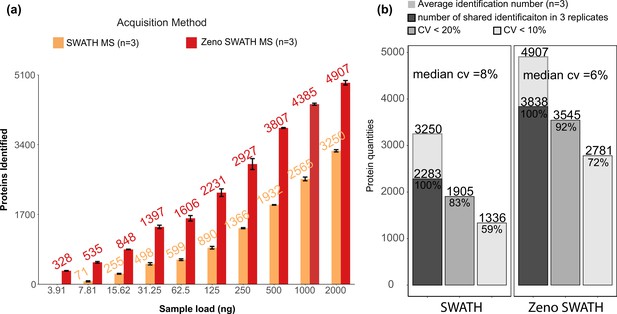
Protein identification performance on K562 human cell line standard using 800 µl/min, 5 min gradient chromatography in SWATH and Zeno SWATH MS.
(a) Comparison of sample injection amount and identification performance with SWATH and Zeno SWATH MS. Illustrated is the average number of proteins identified (n=3, error bars [±1 SD] shown in plot) for a K562 human cell line standard dilution series under two acquisition methods with spectral library DIA-NN analysis. (b) Reproducibility of proteins quantified in SWATH and Zeno SWATH MS on 2 µg of K562 human cell line standard separated by analytical flow-rate chromatography. Average protein identification number in three replicates (background), number of proteins identified in all replicates (dark grey), proteins with coefficient of variation below 20% (grey) and below 10% (light grey) of SWATH and Zeno SWATH MS. Raw data were analysed by spectral library-based DIA-NN analysis.
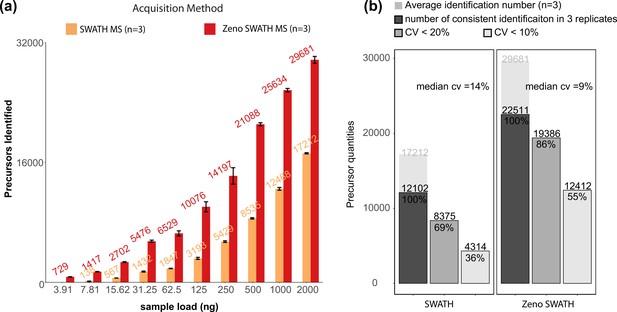
Precursor identification performance on K562 human cell line standard using 800 µl/min, 5 min gradient chromatography in SWATH and Zeno SWATH MS.
(a) Comparison of sample injection amount and precursor identification performance with SWATH and Zeno SWATH MS. Illustrated is the average number of precursors identified (n=3, error bars [±1 SD] shown in plot) for a K562 human cell line standard dilution series under two acquisition methods with spectral library DIA-NN analysis. (b) Reproducibility of precursors quantified in SWATH and Zeno SWATH MS on 2 µg of K562 human cell line standard separated by analytical flow-rate chromatography. Average precursor identification number in three replicates (background), number of precursors identified in all replicates (dark grey), precursors with coefficient of variation below 20% (grey) and below 10% (light grey) of SWATH and Zeno SWATH MS. Raw data were analysed by spectral library-based DIA-NN analysis.
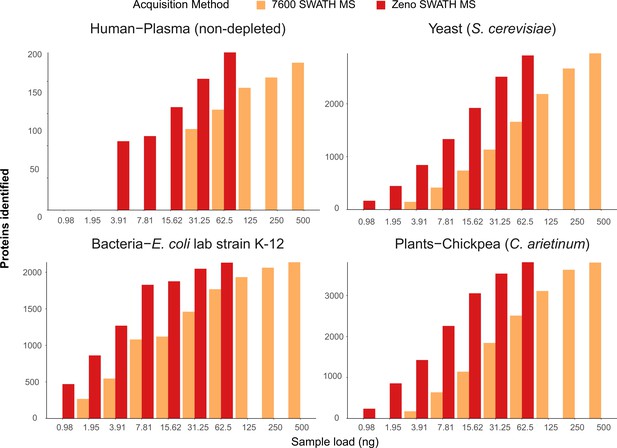
Protein identification number with SWATH MS and Zeno SWATH MS in different sample types with 5 µl/min, 20 min micro-flow-rate chromatography.
We generated tryptic digests from human plasma, and protein extracts from the yeast Saccharomyces cerevisiae S228c, the Escherichia coli lab strain K-12, and chickpea (Cicer arietinum) seedlings germinated in the lab. Data were processed with library-free DIA-NN analysis. Shown data are the identification numbers of and concentration, detailed information of numbers can be found in Supplementary file 1.
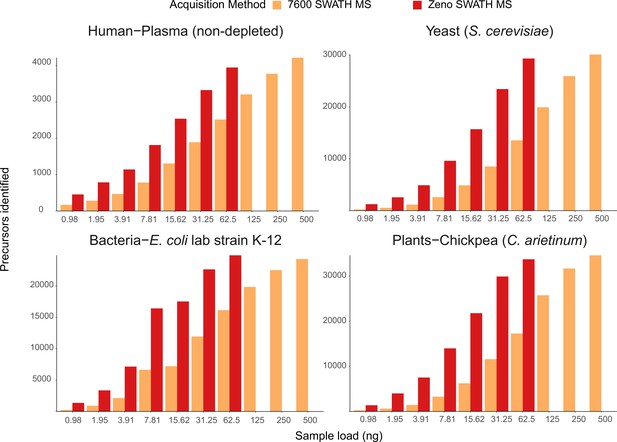
Precursor identification with SWATH acquisition and Zeno SWATH MS in different sample types with 5 µl/min, 20 min micro-flow-rate chromatography.
We generated tryptic digests from human plasma, and protein extracts from the yeast Saccharomyces cerevisiae S288c, the Escherichia coli lab strain K-12, and chickpea (Cicer arietinum) seedlings germinated in the lab. Data were processed with library-free DIA-NN analysis. Shown data are the identification number of respective species and concentration, detailed information of numbers can be found in Supplementary file 1.
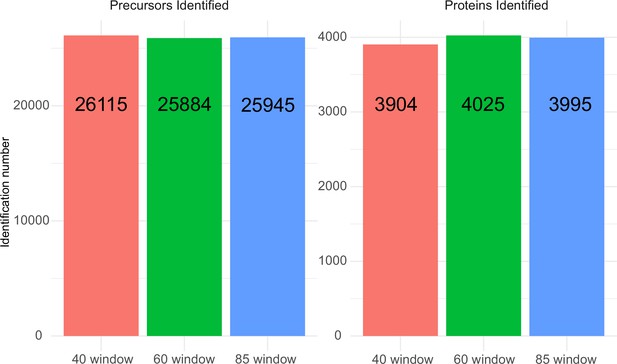
Precursor and protein identification performance with 100 ng K562 human cell line lysate injection using 5 µl/min, 20 min micro-flow chromatography coupled with different SWATH window number acquisition schemes.
For the 40 variable window SWATH acquisition scheme, 26,115 precursors from 3904 proteins were identified; the 60 variable window SWATH acquisition scheme identified 25,884 precursors from 4025 proteins; and the 85 variable window SWATH acquisition scheme identified 25,945 precursors from 3995 proteins.
Additional files
-
Supplementary file 1
Precursors and proteins identification number in all sample species – precursors identification.
- https://cdn.elifesciences.org/articles/83947/elife-83947-supp1-v1.xlsx
-
Supplementary file 2
LC-MS instrumentation parameters.
- https://cdn.elifesciences.org/articles/83947/elife-83947-supp2-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83947/elife-83947-mdarchecklist1-v1.docx





