Dopamine signaling regulates predator-driven changes in Caenorhabditis elegans’ egg laying behavior
Figures
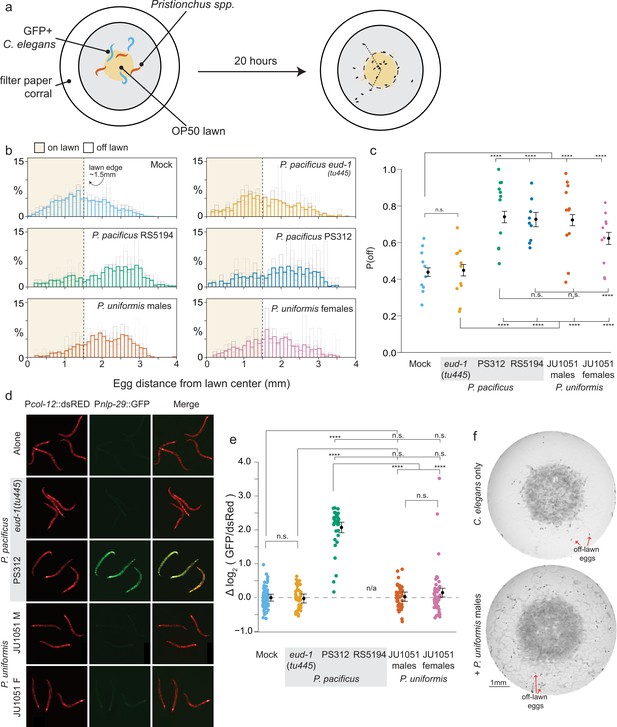
Predators influence prey egg location.
(a) Schematic showing egg location assay setup. Small lawns (approx. 3 mm) in diameter are enclosed in a filter paper corralled arena. Six animals are placed into the arena, three GFP+ C. elegans strain CX7389 and three Pristionchus spp. (or six C. elegans in mock controls). After 20 hr, eggs are visualized and <x,y> positions in the arena are determined. (b) Histograms of egg distributions in mock (N=10 arenas) or five predator conditions: P. pacificus eud-1(tu445) mutants (N=11 arenas), P. pacificus strain PS312 (California isolate) (N=11 arenas), P. pacificus strain RS5194 (Japan isolate) (N=9 arenas), P. uniformis strain JU1051 males (N=11 arenas), and P. uniformis JU1051 females (N=10 arenas). Bolded bars show average distribution of egg distance from center (in mm) with faint bars indicating the individual arena distributions. Lawn edge is marked at radial distance approximately 1.5 mm from center. (c) Distributions of eggs are quantified as <# eggs off lawn, # eggs on lawn> in each arena and the observed probability of off lawn egg laying (P(off)) is plotted in each condition (# eggs off lawn/total # of eggs, average of ~90 eggs per arena). Statistical analysis was performed by logistic regression in R modeling the [# off, # on] egg counts as a function of predator condition, with significant effects determined by likelihood ratio analysis of deviance in R. Model estimates are overlaid on plots as expected values of P(off) from the logistic model ± 95% confidence intervals. We detected a significant main effect of predator condition (p<2.2 × 10–16). Post hoc comparisons with correction for multiple testing were computed using the single step multivariate normal procedure in the multcomp package in R according to simultaneous method of Hothorn, Brez, and Westfall (Hothorn et al., 2008). (d) C. elegans expressing Pnlp-29::GFP and a Pcol-12::dsRed co-injection marker paired with various predators after 20 hr. P. pacificus RS5194 animals all died following 20 hr of predator exposure. GFP signal was quantified and normalized to dsRed signal for each animal. (e) log2 (GFP/dsRed) signal is shown relative to the mock mean (=0). N=79 mock, 47 P. pacificus eud-1(tu445), 34 P. pacificus PS312, 44 P. uniformis JU1051 males, 49 P. uniformis JU1051 females. Statistical analysis was performed with ANOVA and we detected a significant main effect of predator condition (p<2.2 × 10–16). Model estimates are overlaid on plots as mean log2 normalized fluorescence ± 95% confidence intervals. Post hoc comparisons with correction for multiple testing were performed using the single step multivariate t procedure in the multcomp package in R (Hothorn et al., 2008). (f) Representative images of egg location assay plates after 20 hr of mock (upper) or exposure to P. uniformis males (lower). Red arrows indicate example eggs laid off lawn. n.s.=p>0.1, †=p<0.1, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 1—source data 1
Egg position data in various predator conditions.
For each test arena, data tabulate the arena, strain, condition, <x,y> coordinates in pixels, lawn radius in pixels, the egg distance from arena center in pixels, the egg distance from lawn edge in pixels, position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Pnlp-29::GFP and Pcol-12::dsRed data in various predator conditions.
Individual animal fluorescence data is tabulated with strain, predator condition, GFP intensity, dsRed intensity, the calculated GFP/dsRed ratio, the calculated log2 GFP/dsRed ratio, and the normalized log2 GFP/dsRed set relative to the average of the mock (control) condition.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig1-data2-v1.xlsx
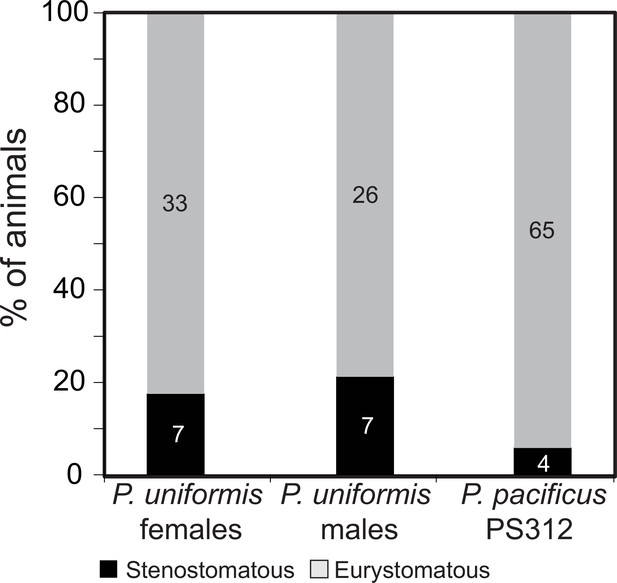
Eurystomatous and stenostomatous animals in P. uniformis and P. pacificus.
Proportion of stenostomatous and eurystomatous animals in P. uniformis females (N=40), P. uniformis males (N=33), and P. pacificus strain PS312 (N=69). We did not detect significant differences in the steno/eurystomatous ratio between P. uniformis males and females (Fisher’s exact test, p=0.77). We did detect a significantly lower proportion of stenostomatous animals in P. pacificus PS312 compared to P. uniformis males (Fisher’s exact test, p=0.036) and a trend to lower proportion of stenostomatous animals compared to P. uniformis females (Fisher’s exact test, p=0.095).

6 hour time course of off lawn egg laying with various predators.
We calculated the number of eggs laid off and on from egg positional data from 1 - 6 hours in either Mock or predator conditions as in Figure 1 ( N=8 - 13 arenas per condition). Numbers of eggs laid off the lawn were analyzed by logistic regression as a two-way interaction model with time as a continuous variable and predator condition. We detected a significant two-way interaction between time and predator condition (likelihood ratio p=3.35 × 10–8). To explore these effects visually, observed P(off) data points are plotted with the trend lines ± 95% confidence intervals from logistic regression. Data points are shown only once per condition, but for reference the Mock control trendline is shown in each panel. All conditions showed significant positive relationship with respect to time indicating P(off) increased over the course of 6 hours. Pairwise comparisons at individual time points between predator and Mock conditions were computed with correction for multiple testing using the single step method in the multcomp package in R as in Figure 1. n.s.=p>0.1, †=p<0.1, * p<0.05, ** p<0.01, ***p<0.001, ****<p<0.0001.
-
Figure 1—figure supplement 2—source data 1
Egg position data in various predator conditions from 1 to 6 hr.
For each test arena, data tabulate the arena, strain, condition, <x,y> coordinates in pixels, lawn radius in pixels, the egg distance from arena center in pixels, the egg distance from lawn edge in pixels, position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm, time in hr.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig1-figsupp2-data1-v1.xlsx
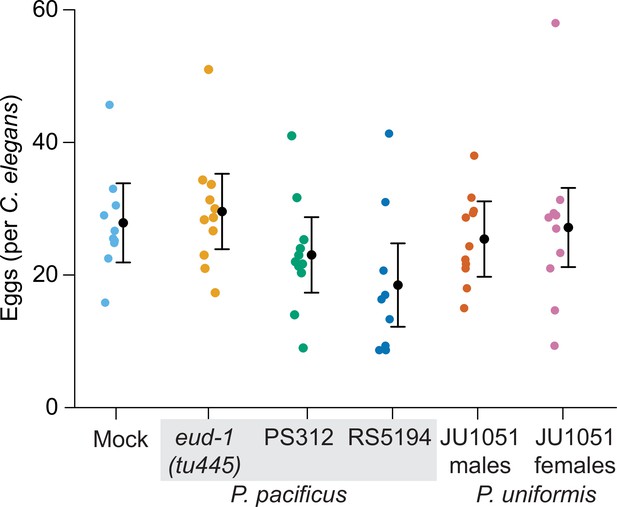
Number of eggs laid in arenas after 20 hr of exposure to various predators.
Average number of eggs per individual C. elegans animal from assays shown in Figure 1B and C are plotted for each predator condition. We did not detect a significant effect of predator condition on the number of eggs laid (one-way ANOVA, p=0.14). Expected values are overlaid on plots as mean number of eggs ± 95% confidence intervals.
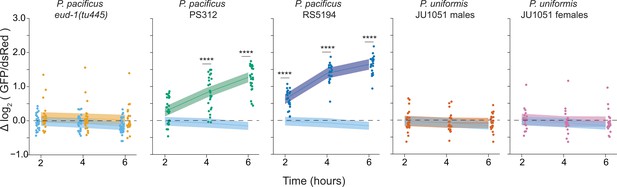
Six hour time course of Pnlp-29::GFP fluorescence with various predators.
We determined the log2 (Pnlp-29::GFP/Pcol-12::dsRed) relative to the mock condition mean from 2 to 6 hr. Relative fluorescence data were analyzed by ANOVA/linear regression as a two-way interaction of time as a continuous variable and predator condition. We detected a significant two-way interaction of predator condition and time (ANOVA p<2.2 × 10–16). To explore this interaction visually, observed fluorescence data are only plotted once per condition, but trendlines ± 95% confidence intervals are shown for each pairwise comparison of predator condition to mock control. In mock conditions, P. uniformis male and P. uniformis female conditions, we did not observe a significant relationship of fluorescence over time, but a positive relationship was observed for both P. pacificus strains. Pairwise comparisons at individual time points between predator and mock conditions were computed with correction for multiple testing using the single step method in the multcomp package in R as in Figure 1. n.s.=p>0.1, †=p<0.1, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 1—figure supplement 4—source data 1
Pnlp-29::GFP and Pcol-12::dsRed data in various predator conditions from 2 to 6 hr.
Individual animal fluorescence data is tabulated with strain, predator condition, GFP intensity, dsRed intensity, the calculated GFP/dsRed ratio, the calculated log2 GFP/dsRed ratio, the normalized log2 GFP/dsRed set relative to the average of the mock (control) condition, and time in hr.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig1-figsupp4-data1-v1.xlsx

Different ratios of predator:prey alter the probability of off lawn egg laying.
(a) The number eggs laid off and on lawn was determined in different ratios of predator:prey (0:6 N=9 arenas, 1:5 N=8 arenas, 2:4, 3:3, 4:2, 5:1 all N=10 arenas). Plotted are observed P(off) in each ratio condition. <# off, #on>data were analyzed by logistic regression/analysis of deviance. Overlaid are model expected values of P(off)±95% confidence intervals. We detected a significant main effect of predator:prey ratio (likelihood ratio p<2.2 × 10–16). Post hoc comparisons with correction for multiple testing were computed using the single step method in the multcomp package in R. (b) The average number of eggs per individual C. elegans in the same arenas as (a) was calculated and data analyzed by ANOVA. Overlaid are mean values ± 95% confidence intervals. We did not detect a significant effect of predator:prey ratio on the number of eggs laid (ANOVA p=0.92). n.s.=p>0.1, †=p<0.1, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 1—figure supplement 5—source data 1
Egg position data in various predator:prey ratios.
For each test arena, data tabulate the arena, ratio of predator:prey, <x,y> coordinates in pixels, lawn radius in pixels, the egg distance from arena center in pixels, the egg distance from lawn edge in pixels, position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig1-figsupp5-data1-v1.xlsx
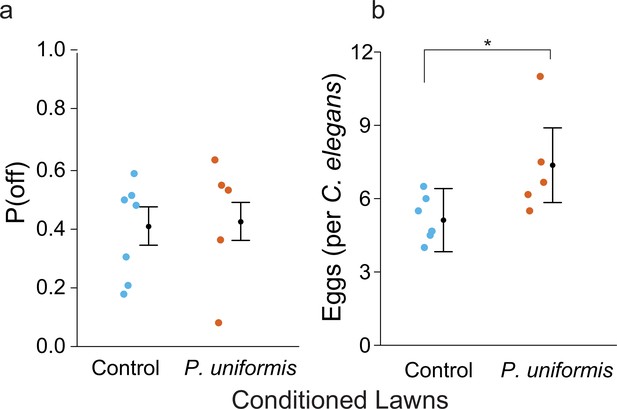
Bacteria pre-conditioned with P. uniformis males is not sufficient to alter egg laying behavior.
(a) The number of eggs laid off and on lawn was determined in arenas with lawns pre-conditioned with mock (sterile C. elegans) (N=7 arenas) or P. uniformis males (N=5 arenas) with six C. elegans per arena allowed to lay eggs for 2 hr. Plotted are observed values of P(off) with data further analyzed by logistic regression/analysis of deviance as in Figure 1 and Figure 1—figure supplement 2. Overlaid are logistic model estimates of the expected values of P(off)±95% confidence intervals. We did not detect a significant effect of lawn condition on P(off) (likelihood ratio p=0.73). (b) The average number of eggs per individual C. elegans animal in arenas in (a) was determined and is plotted for each condition, with data analyzed by ANOVA. Overlaid are mean estimates ± 95% confidence intervals. We did detect a significant increase to the average number of eggs laid in lawns conditioned by P. uniformis males (ANOVA p=0.03). *p<0.05.
-
Figure 1—figure supplement 6—source data 1
Egg position data in arenas with conditioned lawns.
For each test arena, data tabulate the arena, condition, <x,y> coordinates in pixels, lawn radius in pixels, the egg distance from arena center in pixels, the egg distance from lawn edge in pixels, position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig1-figsupp6-data1-v1.xlsx
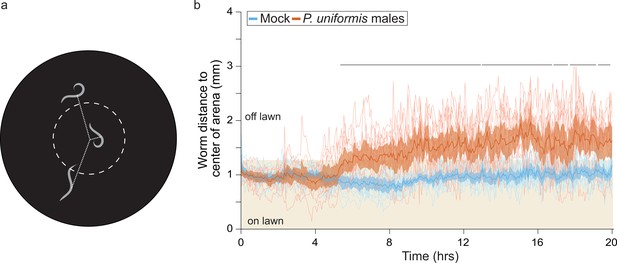
C. elegans shows sustained avoidance of the lawn when exposed to predator.
(a) Schematic for WormWatcher experiments for location tracking. Distance of midpoint of fluorescent C. elegans (strain ARM112, Peft-3::mScarlet) to center of the arena is tracked over 20 hr (15 frames per hour, tresolution = 4 min). (b) Worms tracked by WormWatcher included ARM112 strain C. elegans in mock (N=12 arenas), or predator (P. uniformis males, N=12 arenas), and are plotted as individual traces (thin lines, average position of all worms in an arena, range 2–6 worms per arena, average = 3), representing average distance from center in mm over time. Data were analyzed by non-parametric bootstrap resampling with replacement with 1×105 iterations. Bold lines represent the estimated average distance over time, with shading representing empirically determined 2.5–97.5% quantiles (95% interval) of bootstrap samples. p<0.05 significance can be inferred from regions of lack of overlap of bootstrapped intervals between mock and predator-exposed conditions, identified with lines above traces showing regions of 0% overlap. Regions with 0% overlap account for 71% of all time points, all occurring in the region >5 hr.
-
Figure 2—source data 1
WormWatcher tracking data for predator and mock-exposed ARM112 mScarlet expressing C. elegans.
For each test arena, 20 hr (15 frames/hr, tresolution = 4 min) of tracking data are tabulated, showing frame number, time (in hr), arena, condition, number of worms tracked per time point, lawn radius (in pixels), distance of body midpoint to center of arena (in pixels), distance of body midpoint to edge of lawn (in pixels), position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig2-data1-v1.xlsx
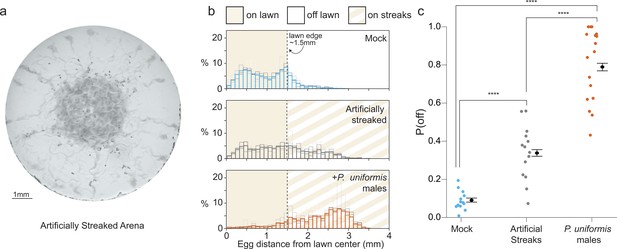
Bacterial topography alone does not account for predator associated changed to egg laying location.
(a) Representative image of an assay plate after 20 hr with artificially streaked lawns. (b) Histograms of egg distributions in mock (N=14 arenas), artificially streaked (N=14 arenas), and predator-exposed (N=17 arenas) conditions. Bolded bars show average distribution of egg distance from center (in mm) with faint bars indicating the individual arena distributions. Lawn edge is marked at radial distance approximately 1.5 mm from center. (c) Distributions of eggs are quantified as [# off lawn, # on lawn] in each arena as in Figure 1, and the observed probability of off lawn laying (P(off)) is plotted in each condition, with data analyzed by logistic regression/analysis of deviance. Overlaid are logistic model estimates of the expected values of P(off)±95% confidence intervals. We detected a significant effect of condition (likelihood ratio p<2.2 × 10–16). Post hoc comparisons with correction for multiple testing were computed using the single step method in the multcomp package in R. n.s.=p>0.1, †=p<0.1, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 3—source data 1
Egg position data in arenas with and without predator exposure and artificial streaking.
For each test arena, data tabulate the arena, condition, <x,y> coordinates in pixels, lawn radius in pixels, the egg distance from arena center in pixels, the egg distance from lawn edge in pixels, position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig3-data1-v1.xlsx
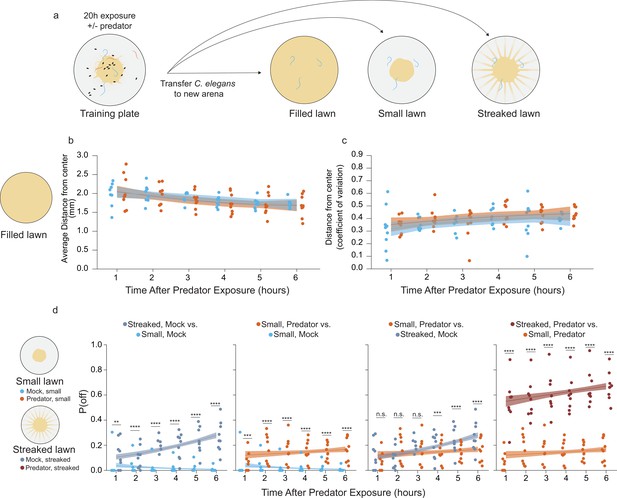
Sustained changes to egg laying is observed following prior predator exposure.
(a) Schematic of egg laying learning assay: after 20 hr of exposure to either mock (C. elegans only) or predator condition (P. uniformis males), worms are transferred to arenas either completely filled with bacteria or arenas with a normal sized small lawn or a lawn with artificial streaks. In the predator-exposed condition, all three C. elegans are transferred, while in the mock condition, three C. elegans selected at random from among the six are transferred. (b) Analysis of distributional properties of C. elegans egg laying for 1–6 hr in arenas completely filled with bacteria after mock (N=8–9 arenas per time point), or predator exposure (N=9 arenas per time point). Plotted are the mean distance from lawn center in mm for each time point and condition. (c) Data points represent the coefficient of variation (standard deviation divided by the mean) for egg distances in (b) for each time point and for each condition. Data in (b) and (c) were analyzed by linear regression/ANOVA modeling interactions of time as continuous variable and predator exposure condition. Overlaid on plots are trendlines for each condition from linear models with shading showing 95% confidence intervals. We detected a significant main effect of time on both average distance from center (ANOVA p=3.0 × 10–6) as well as on the dispersal of the eggs measured by the coefficient of variation (ANOVA p=0.0016) but no significant main effect of predator condition (average distance, p=0.51; coefficient of variation, p=0.14) or interaction effects on either variable (average distance, p=0.76; coefficient of variation, p=0.97). (d) Analysis of off lawn egg laying in animals exploring small lawns or lawns with artificial streaks after 20 hr of mock or predator exposure. Data points in (d) represent observed P(off) in each time point and condition (N=9–12 arenas per time point/condition). Off lawn egg laying probability was analyzed by logistic regression/analysis of deviance modeling a three-way interaction between time as a continuous variable, lawn type, and predator exposure condition. We detected a significant three-way interaction between these independent variables (likelihood ratio p=1.5 × 10–7). Data in D–G were analyzed together as a single analysis paradigm, however to ease visual understanding of this interaction, pairwise comparisons between conditions are shown in separate panels D–G for: artificially streaked and small lawns for mock-exposed animals, predator vs. mock in small lawns, predator exposure/small lawns compared to the artificially streaked/mock-exposed animals, and finally artificially streaked lawns compared to small lawns for predator-exposed animals. Pairwise comparisons at individual time points between lawn types/conditions were computed with correction for multiple testing using the single step method in the multcomp package in R. n.s.=p>0.1, †=p<0.1, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 4—source data 1
Egg position data in filled arenas after predator exposure.
For each test arena, data tabulate the arena, condition, <x,y> coordinates in pixels, the egg distance from arena center in pixels, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, time in hr.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Egg position data in small or artificially streaked arenas after predator exposure.
For each test arena, data tabulate the arena, predator exposure condition, lawn type, <x,y> coordinates in pixels, lawn radius in pixels, the egg distance from arena center in pixels, the egg distance from lawn edge in pixels, position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm, time in hr.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig4-data2-v1.xlsx
-
Figure 4—source data 3
Table of slopes for temporal changes in the distributional properties of eggs after predator exposure in filled arenas.
Slopes showing the model estimates for temporal change per hour in mean distance from center (mm/hr) and coefficient of variation (units/hr) in Figure 4b and c in predator-exposed and control conditions.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig4-data3-v1.xlsx
-
Figure 4—source data 4
Table of slope for temporal changes to the probability of off lawn egg laying with and without predator exposure, and in arenas with differing bacterial topology.
Slopes showing the model estimates for temporal change per hour in log odds ratio () from logistic models in Figure 4d in different conditions. On logit scale, a slope of +0.1 = 1.1-fold, 0.22=1.25-fold, +0.4 = 1.5-fold, +0.69 = 2-fold change in the P(off)/P(on) odds ratio.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig4-data4-v1.xlsx
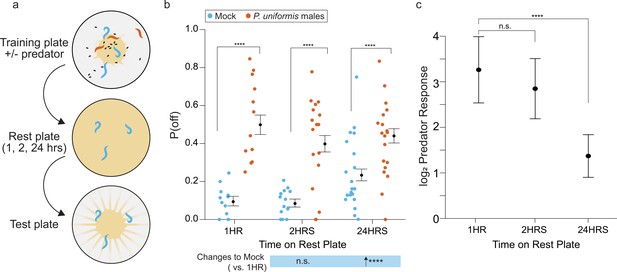
Changes to egg laying behavior after predator exposure continue for 24 hr.
(a) Schematic of egg laying learning assay. C. elegans are exposed to mock or predator condition (P. uniformis males) for 20 hr and transferred to a rest plate for 1, 2, or 24 hr. After rest, animals are transferred to a test arena containing artificial streaks as in Figure 4 and positions of laid eggs are determined in order to determine the proportion of eggs laid off and on the lawn. (b) Observed P(off) in test arenas is plotted by condition and length of rest period (mock/1 hr N=12 arenas, mock/2 hr N=15 arenas, mock/24 hr N=20 arenas, predator-exposed/1 hr N=12 arenas, predator-exposed/2 hr N=17 arenas, predator-exposed/24 hr N=19 arenas). Data were analyzed by logistic regression/analysis of deviance fitting a two-way interaction of categorical length of rest period (1–24 hr) and mock or predator condition, with expected values of P(off)±95% confidence intervals from logistic model overlaid on plot. We found a significant two-way interaction of rest period length and predator exposure condition (likelihood ratio p=3.4 × 10–11). (c) Log2 fold change in computed predator response is plotted for each rest time period, where predator response is defined as the change to the odds ratio of [off lawn/on lawn] egg laying between predator and mock conditions (see Materials and methods, Equations 1–3). These are displayed as point estimates with 95% confidence intervals as derived from logistic regression. Post hoc comparisons between conditions, as well as changes to predator response, with correction for multiple testing, were computed using the single step method in the multcomp package in R as in previous figures. n.s.=p>0.1, †=p<0.1, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 5—source data 1
Egg position data after periods of 1 hr, 2 hr, or 24 hr following predator exposure.
For each test arena, data tabulate the arena, predator exposure condition, rest period, <x,y> coordinates in pixels, lawn radius in pixels, the egg distance from arena center in pixels, the egg distance from lawn edge in pixels, position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig5-data1-v1.xlsx
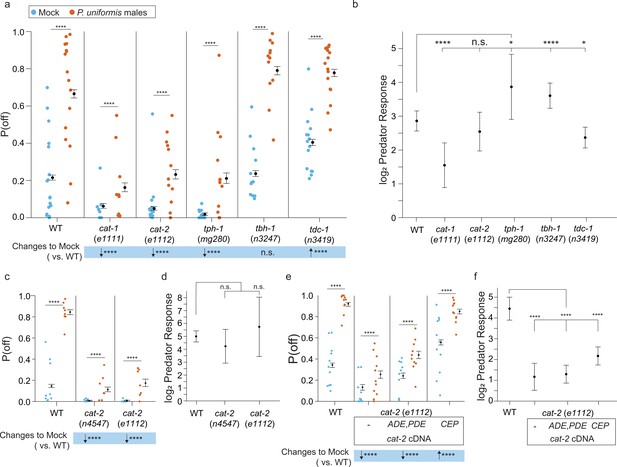
Loss of biogenic amine synthesis results in changes to the probability of laying eggs off the bacterial lawn.
(a) Plotted are observed P(off) data for either mock or predator-exposed arenas in various mutants in biogenic amine synthesis genes (mock: wildtype [WT] N=17 arenas, cat-1(e1111) N=18, cat-2(e1112) N=13, tph-1(mg280) N=12, tbh-1(n3247) N=12, tdc-1(n3419) N=15, predator-exposed: WT N=16, cat-1(e1111) N=9, cat-2(e1112) N=12, tph-1(mg280) N=12, tbh-1(n3247) N=12, tdc-1(n3419) N=16). Data were analyzed by logistic regression/analysis of deviance fitting a two-way interaction of genotype and predator exposure, with overlaid expected values of P(off) from logistic modeling±95% confidence intervals. We detected a significant two-way interaction of genotype and predator exposure condition (likelihood ratio p<2.2 × 10–16). (b) Log2 predator response (as in Figure 5 and Materials and methods, Equation 3) is plotted as point estimates with error bars showing 95% confidence intervals across genotypes. (c) Observed P(off) data in mock or predator-exposed conditions in WT or two cat-2 mutant alleles n4547 and e1112 (mock N=9 arenas per genotype, predator N=8 arenas per genotype). Data analyzed as in (a) with overlaid expected values for P(off) from the logistic model±95% confidence intervals. We failed to detect a significant interaction between genotype and predator condition (likelihood ratio p=0.22) but we were able to detect a main effect of genotype (p<2.2 × 10–6) and a main effect of predator exposure (p<2.2 × 10–6). (d) Log2 predator response across genotypes as in (b). (e) Observed P(off) in WT or cat-2(e1112) mutant animals with or without transgenic rescue of cat-2 cDNA in either ADE/PDE or CEP neurons (mock/WT N=11 arenas, predator/WT N=10 arenas, mock/cat-2(e1112) N=10 arenas, predator/cat-2(e1112) N=11 arenas, cat-2(e1112); p27::cat-2-sl2-GFP (ADE/PDE) N=10 arenas for each condition, cat-2(e1112); Pdat-1p19::cat-2-sl2-GFP (CEP) N=11 arenas per condition). Data analyzed as in (a, c) with overlaid expected values for P(off) from the logistic model±95% confidence intervals. We detected a significant two-way interaction of genotype and predator exposure condition (likelihood ratio p<2.2 × 10–16). (f) Log2 predator response as described in (b) and (d) across genotype/transgenic rescue conditions. Post hoc with correction for multiple testing, were computed using the single step method in the multcomp package in R as in previous figures. n.s.=p>0.1, †=p<0.1, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 6—source data 1
Egg position data in biogenic amine mutants with and without predator exposure.
For each test arena, data tabulate the arena, predator exposure condition, genotype, <x,y> coordinates in pixels, lawn radius in pixels, the egg distance from arena center in pixels, the egg distance from lawn edge in pixels, position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig6-data1-v1.xlsx
-
Figure 6—source data 2
Egg position data in cat-2 mutant alleles with and without predator exposure.
For each test arena, data tabulate the arena, predator exposure condition, genotype, <x,y> coordinates in pixels, lawn radius in pixels, the egg distance from arena center in pixels, the egg distance from lawn edge in pixels, position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig6-data2-v1.xlsx
-
Figure 6—source data 3
Egg position data in cat-2 mutant alleles with and without predator exposure and rescue of cat-2 cDNA.
For each test arena, data tabulate the arena, predator exposure condition, genotype, <x,y> coordinates in pixels, lawn radius in pixels, the egg distance from arena center in pixels, the egg distance from lawn edge in pixels, position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig6-data3-v1.xlsx

Mutants in dopamine synthesis and reuptake show varying degrees of predator avoidance.
(a) WormWatcher data tracking position (15 frames per hour, tresolution = 4 min) in ARM112 mScarlet-expressing C. elegans (‘wildtype [WT]’) exposed to mock (N=8 arenas) or predator (N=8 arenas) over the course of 20 hr. (b) Position tracking of ARM112 animals crossed into the cat-2(e1112) mutant background in mock (N=8) or predator (N=9) conditions. Data in (a, b) are plotted as individual traces (thin lines) representing average distance from center over time per arena. Data were analyzed by non-parametric bootstrap resampling with replacement with 1×105 iterations. Bold lines represent the estimated average distance over time, with shading representing empirically determined 2.5–97.5% quantiles (95% interval) of bootstrap samples. p<0.05 significance can be inferred from regions of lack of overlap of bootstrapped intervals between mock and predator-exposed conditions, identified with lines above traces showing regions of 0% overlap, accounting for 63% of all time points in (a) and 38% of time points in (b). (c) The bootstrap estimates for the magnitude of change between predator-mock distance from center in both genotypes. (d, e) WormWatcher data tracking position as in (a, b) in either WT ARM112 controls or ARM112 crossed into the dat-1(ok157) mutant background. Lack of bootstrap confidence interval overlap between conditions is evident for 76% of all time points for WT and 82% of all time points for dat-1(ok157) mutants. (f) Bootstrap estimates for the magnitude of change between predator-mock distance from center in both genotypes.
-
Figure 6—figure supplement 1—source data 1
WormWatcher tracking data for predator and mock-exposed ARM112 mScarlet expressing C. elegans and ARM112 animals with cat-2(e1112) mutant allele.
For each test arena, 20 hr (15 frames/hr, tresolution = 4 min) of tracking data are tabulated, showing frame number, time (in hr), arena, condition, genotype, number of worms tracked per time point, lawn radius (in pixels), distance of body midpoint to center of arena (in pixels), distance of body midpoint to edge of lawn (in pixels), position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig6-figsupp1-data1-v1.xlsx
-
Figure 6—figure supplement 1—source data 2
WormWatcher tracking data for predator and mock-exposed ARM112 mScarlet expressing C. elegans and ARM112 animals with dat-1(ok157) mutant allele.
For each test arena, 20 hr (15 frames/hr, tresolution = 4 min) of tracking data are tabulated, showing frame number, time (in hr), arena, condition, genotype, number of worms tracked per time point, lawn radius (in pixels), distance of body midpoint to center of arena (in pixels), distance of body midpoint to edge of lawn (in pixels), position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig6-figsupp1-data2-v1.xlsx
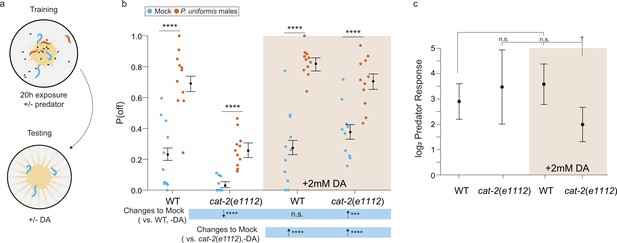
Addition of exogenous dopamine rescues egg laying behavior in dopamine synthesis deficient mutants.
(a) Schematic of the egg laying learning assay. C. elegans exposed to either mock or predator condition for 20 hr are transferred to testing arenas containing artificially streaked bacteria with or without the addition of 2 mM dopamine. (b) Observed P(off) data are plotted for each genotype, predator exposure, and presence of exogenous dopamine (N=11 arenas all conditions except cat-2(e1112)/mock/+3 mM dopamine condition which had N=10 arenas). Data were analyzed as in previous figures by logistic regression/analysis of deviance fitting a three-way interaction of genotype, predator exposure, and dopamine, with overlaid expected values of P(off) from logistic modeling±95% confidence intervals. We detected a significant three-way interaction of genotype, predator exposure, and dopamine (likelihood ratio p=0.0006). (c) Log2 predator response±95% confidence intervals (as in Figures 5–6, see Materials and methods, Equation 3) in each genotype with and without addition of 2 mM dopamine. Post hoc with correction for multiple testing were computed using the single step method in the multcomp package in R as in previous figures. n.s.=p>0.1, †=p<0.1, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 7—source data 1
Egg position data in cat-2(e1112) mutants with and without predator exposure and addition of 3 mM dopamine.
For each test arena, data tabulate the arena, predator exposure condition, genotype, dopamine addition, <x,y> coordinates in pixels, lawn radius in pixels, the egg distance from arena center in pixels, the egg distance from lawn edge in pixels, position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig7-data1-v1.xlsx
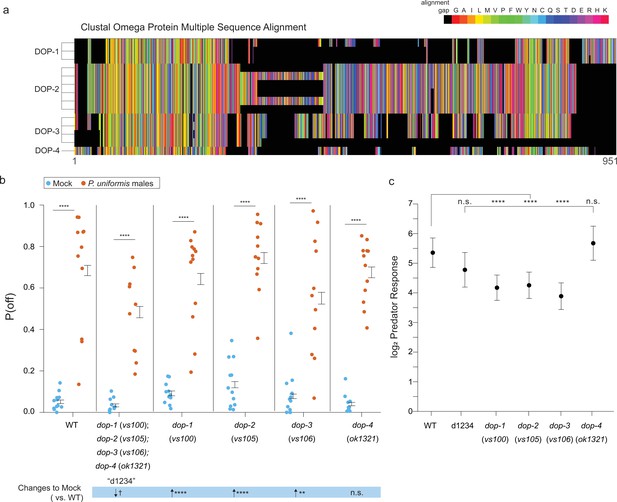
Mutations in DOP family dopaminergic receptors influence egg laying behavior with predator exposure.
(a) CLUSTAL Omega multiple protein sequence alignment of the three isoforms of dopaminergic receptors DOP-1, the six of DOP-2, the three of DOP-3, and DOP-4 are shown visually as a colormap where black squares represent sequence alignment gaps, and amino acids colors are grouped by type (e.g. uncharged, charged). (b) Observed P(off) data are shown for the mock and predator-exposed conditions in WT (mock N=12 arenas, predator N=11 arenas), a quadruple mutant for all four receptor genes (N=10/condition), and single receptor mutants dop-1(vs100) (N=12/condition), dop-2(vs105) (mock N=12, predator N=11), dop-3(vs106) (mock N=12, predator N=11), and dop-4(ok1321) (mock N=11, predator N=12). Data were analyzed as in previous figures by logistic regression/analysis of deviance fitting a two-way interaction of genotype and predator exposure, with overlaid expected values of P(off) from logistic modeling±95% confidence intervals. We detected a significant two-way interaction of genotype and predator condition (likelihood ratio p<2.2 × 10–6). (c) Log2 predator response±95% confidence intervals as in Figures 5—7 (see Materials and methods, Equation 3) across genotypes. Post hoc comparisons with correction for multiple testing were computed using the single step method in the multcomp package in R as in previous figures. n.s.=p>0.1, †=p<0.1, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 8—source data 1
CLUSTAL multiple protein sequence alignment of DOP receptor amino acid sequences.
FASTA format for amino acid sequences of DOP-1, DOP-2, DOP-3, DOP-4 proteins, and isoforms obtained from Wormbase.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig8-data1-v1.zip
-
Figure 8—source data 2
Egg position data in dopamine receptor mutants with and without predator exposure.
For each test arena, data tabulate the arena, predator exposure condition, genotype, <x,y> coordinates in pixels, lawn radius in pixels, the egg distance from arena center in pixels, the egg distance from lawn edge in pixels, position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig8-data2-v1.xlsx
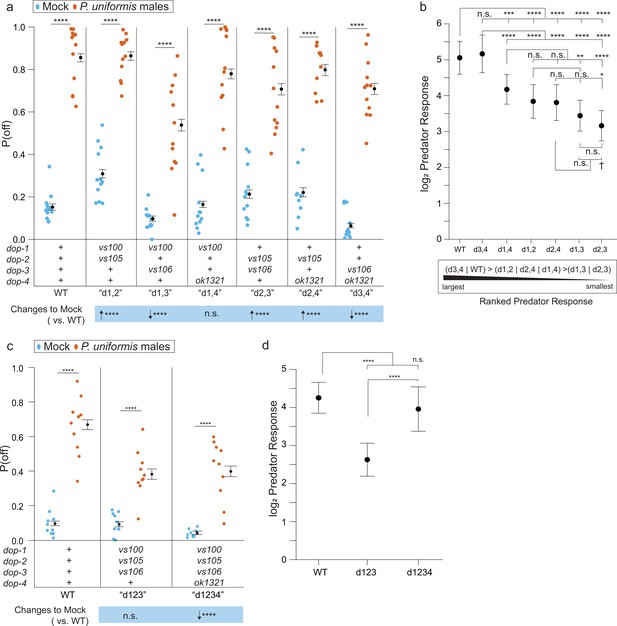
Loss of dopaminergic signaling via combinations of DOP receptors is associated with changes to both baseline egg laying behavior and the magnitude of predator response.
(a) Observed P(off) data are shown for mock and predator-exposed conditions in WT or various pairwise combinations of dopamine receptor mutants (N=12 arenas per condition except mock/dop-2(vs105);dop-4(ok1321) N=9, and predator/dop-2(vs105);dop-4(ok1321) N=10). Data were analyzed as in previous figures by logistic regression/analysis of deviance fitting a two-way interaction of genotype and predator exposure, with overlaid expected values of P(off) from logistic modeling±95% confidence intervals. We detected a significant two-way interaction of genotype and predator condition (likelihood ratio p<2.2 × 10-6). (b) Log2 predator response±95% confidence intervals as in Figures 5—8 (see Materials and methods, Equation 3) across receptor mutant combinations. Below (b) is shown a qualitative visualization of the predator response ranked from highest to lowest. (c) Observed P(off) data shown for mock and predator-exposed conditions in WT, a triple mutant dop-1(vs100);dop-2(vs105);dop-3(vs106), and a quadruple mutant in all four dop- genes (N=10 arenas per condition except mock/quadruple mutant N=9). Data analyzed as in (a) with overlaid expected values for P(off)±95% confidence intervals. We detected a significant two-way interaction of genotype and predator condition (likelihood ratio p=2 × 10–13). (d) Log2 predator response±95% confidence intervals as in (b). Post hoc comparisons with correction for multiple testing were computed using the single step method in the multcomp package in R as in previous figures. n.s.=p>0.1, †=p<0.1, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 9—source data 1
Egg position data in pairwise combinations of dopamine receptor mutants with and without predator exposure.
For each test arena, data tabulate the arena, predator exposure condition, genotype, <x,y> coordinates in pixels, lawn radius in pixels, the egg distance from arena center in pixels, the egg distance from lawn edge in pixels, position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig9-data1-v1.xlsx
-
Figure 9—source data 2
Egg position data in triple and quadruple dopamine receptor mutants with and without predator exposure.
For each test arena, data tabulate the arena, predator exposure condition, genotype, <x,y> coordinates in pixels, lawn radius in pixels, the egg distance from arena center in pixels, the egg distance from lawn edge in pixels, position as 1 (off) or 0 (on) lawn, the conversion factor for pixel data in mm-per-pixel, the calculated distance from center in mm, the calculated distance from the lawn edge in mm.
- https://cdn.elifesciences.org/articles/83957/elife-83957-fig9-data2-v1.xlsx
Videos
C. elegans avoids P. uniformis.
Video showing P. uniformis and C. elegans on a bacterial lawn. C. elegans shows rapid avoidance responses to bites from P. uniformis.





