Nucleotide binding is the critical regulator of ABCG2 conformational transitions
Figures
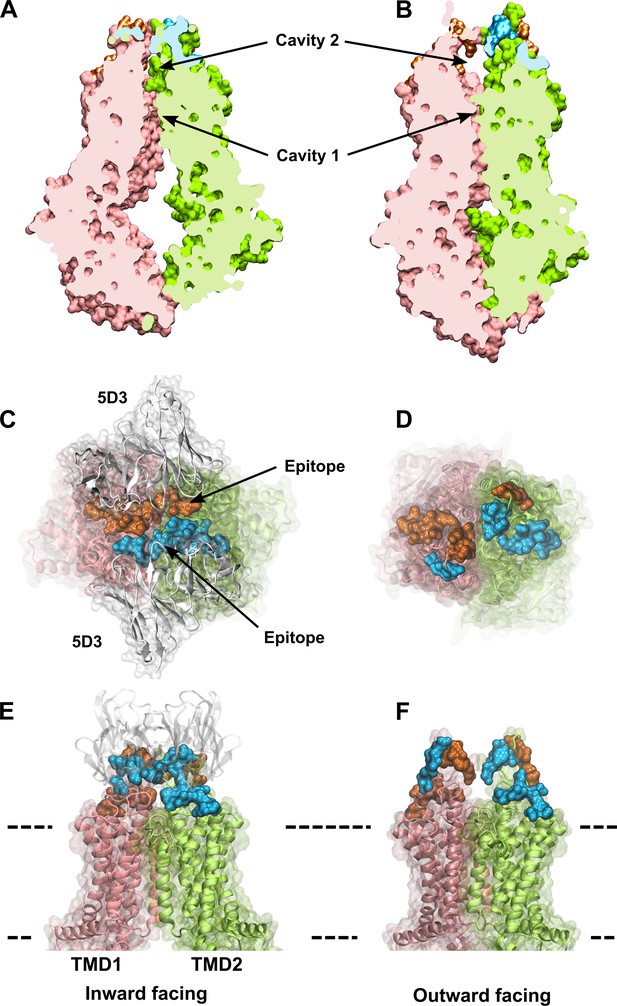
The IF and OF conformers of ABCG2.
A slice through IF (A) and OF (B) ABCG2 shows the lower and upper cavity and reveal their volume changes induced by the transition. Two rotationally symmetric 5D3 epitopes are formed in the IF conformation (C, E) and are absent in the OF conformation of ABCG2 (D, F). Panels (C) and (D) show an extracellular view of the 5D3-bound IF (PDB ID: 6ETI), and the OF (PDB ID: 6HBU) ABCG2 conformations, respectively. A side view of the IF (E) and the OF (F) ABCG2 is also shown. The two protomers of ABCG2 are colored in pink and green in cartoon backbone and transparent surface rendering. The two epitopes are highlighted in blue and orange, and include all residues that are in direct contact with the 5D3 Fab (white) in the 6ETI cryo-EM structure. The membrane boundaries are indicated by dashed lines. cryo-EM, cryogenic electron microscopy; IF, inward-facing; OF, outward-facing.
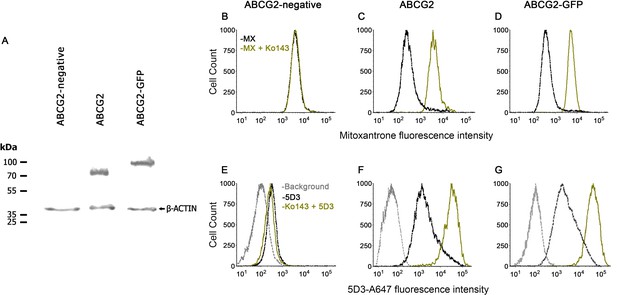
Functional characterization of human ABCG2 and ABCG2-GFP.
Western blot showing comparable expression of ABCG2 and ABCG2-GFP in MDCK II cells using the BXP-21 anti-ABCG2 mAb (A). ABCG2 and ABCG2-GFP expressing cells show decreased mitoxantrone (MX) accumulation, which is reversed to the level of the ABCG2-negative cells by 2 µM Ko143 (B–D). 2 µM Ko143 pre-treatment causes an increase in the 5D3-reactivity of ABCG2 (F) and ABCG2-GFP (G) expressing cells. ABCG2-negative cells show only a negligible 5D3 binding that is not affected by Ko143 (E).
-
Figure 2—source data 1
Source Data to Figure 2A.
- https://cdn.elifesciences.org/articles/83976/elife-83976-fig2-data1-v1.xlsx
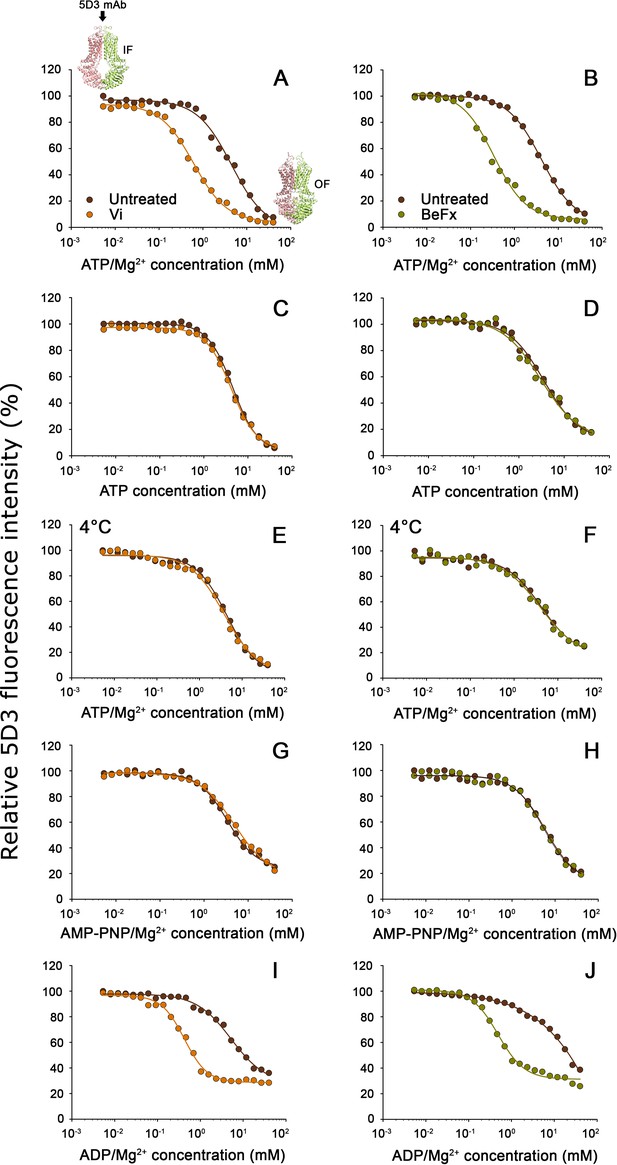
Nucleotide dependence of 5D3-reactivity.
MDCK-ABCG2 cells were permeabilized to allow the titration of intracellular nucleotide concentrations. Dose-response curves of 5D3 binding with increasing concentrations of ATP/Mg2+ (panels (A), and (B)), ATP in the presence of EDTA (panels (C) and (D)), AMP-PNP/Mg2+ (panels (G) and (H)), or ADP/Mg2+ (panels (I) and (J)) were obtained in the presence or absence of Vi (left panels) or BeFx (right panels). Samples were pre-treated with nucleotides at 37°C for 10 min, then further incubated with 5 µg/ml 5D3-A647 at 37°C for 20 min except for (panels (E) and (F)), where all the treatments were carried out on ice. In case of nucleotide trapping, permeabilized cells were co-treated with nucleotides and BeFx or Vi at 37°C for 30 min, then un-trapped nucleotides were washed out and 5D3 labeling was carried out on ice for 45 min. Representative curves are shown from three to five independent experiments. The small inserts in panel (A) depict the IF and OF conformations of ABCG2. IF, inward-facing; OF, outward-facing.
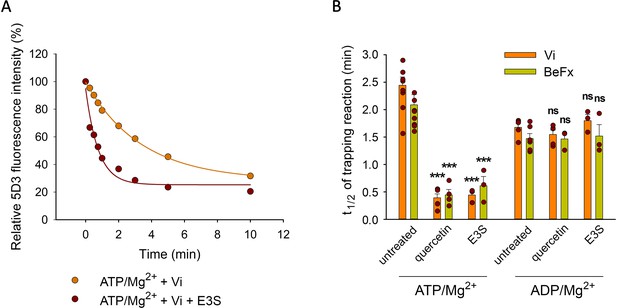
Effect of transported substrates on the kinetics of the formation of the Vi- and BeFx-trapped complexes.
Permeabilized MDCK-ABCG2 cells were pre-treated or not with 10 µM quercetin or E3S for 10 min at 37°C and then further incubated with 0.5 mM ATP/Mg2+ or ADP/Mg2+ in the presence of Vi or BeFx. Samples taken at different time points were stained with 5 µg/ml 5D3-A647 on ice for 45 min. Panel (A) shows a representative Vi-trapping experiment in the absence or presence of E3S, while panel (B) summarizes the t1/2 values calculated from the exponential fit of the kinetic curves (see Materials and methods). Mean ± SD of 3–5 independent experiments is shown. Significant differences compared to substrate untreated samples are shown by ***: p < 0.001.
-
Figure 4—source data 1
Source Data to Figure 4B.
- https://cdn.elifesciences.org/articles/83976/elife-83976-fig4-data1-v1.xlsx
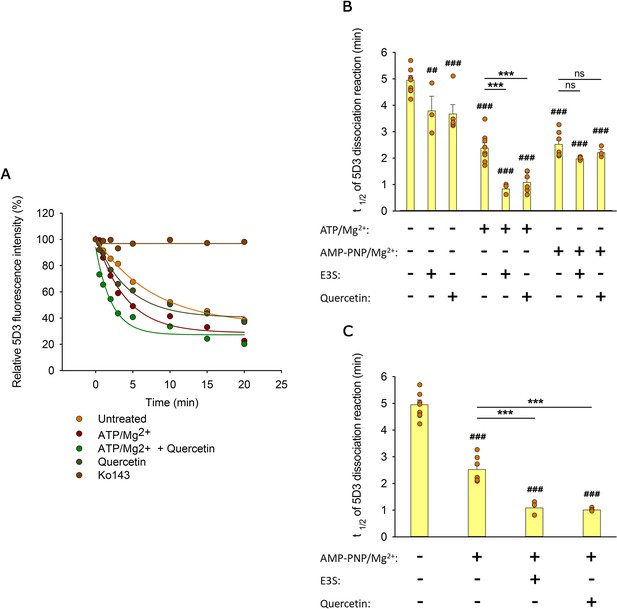
Effects of substrates and nucleotides on the kinetics of 5D3 dissociation when substrates were added simultaneously with nucleotides (A, B), or prior to nucleotides (C).
Permeabilized MDCK-ABCG2 cells were pre-labeled with 5D3-A647 in the absence (panels (A) and (B)) or presence (panel (C)) of substrates (10 µM quercetin or 10 µM E3S) for 20 min at 37°C. After removal of the unbound antibody, cells were further incubated with 3 mM ATP/Mg2+ or AMP-PNP/Mg2+ in the absence or presence of substrates. Panel (A) shows representative 5D3 dissociation curves in the indicated conditions, while panels (B) and (C) summarizes the t1/2 values calculated from the exponential fit of the dissociation curves (see Materials and methods). Means ± SD of 3–5 independent experiments are shown. Significant differences compared to untreated samples (first column) are shown by ###: p < 0.001 or ##: p < 0.01. Significant differences compared to only nucleotide-treated samples are indicated by ***: p < 0.001.
-
Figure 5—source data 1
Source Data to Figure 5B and C.
- https://cdn.elifesciences.org/articles/83976/elife-83976-fig5-data1-v1.xlsx
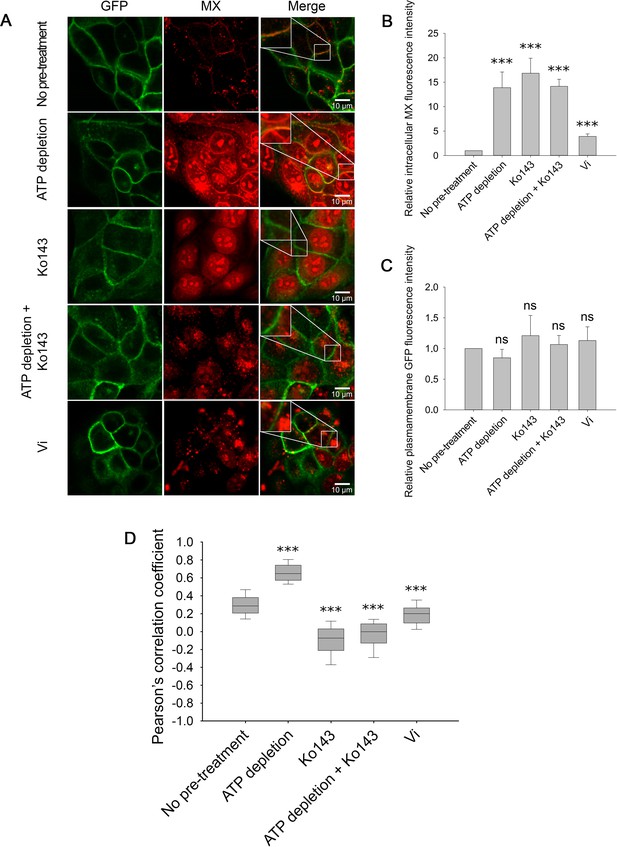
Cellular distribution of MX (red) in MDCK cells expressing ABCG2-GFP (green) in response to ATP depletion and/or Ko143 or Vi treatment (A).
All treatments increased the intracellular MX fluorescence intensity (B), while the ABCG2-GFP fluorescence intensity in the plasma membrane remained unchanged (C). Pearson’s correlation coefficients between the intensity distributions of MX and ABCG2-GFP in the plasma membrane pixels reveal the stabilization of a high-affinity substrate binding ABCG2 conformation in energy-deprived cells (D). ATP depletion was induced by 15 min pre-treatment of cells with 10 mM sodium azide and 8 mM 2-deoxy-D-glucose. Ko143, a non-fluorescent, competitive ABCG2 inhibitor, was added 10 min before MX staining. In panels (B) and (C), bars represent mean ± SD values, while panel (D) shows box and whisker plots. For each treatment group, 150–200 cells were analyzed from three to five independent experiments. Significant differences compared to the untreated control are shown by ***: p < 0.001.
-
Figure 6—source data 1
Source Data to Figure 6B–D.
- https://cdn.elifesciences.org/articles/83976/elife-83976-fig6-data1-v1.xlsx
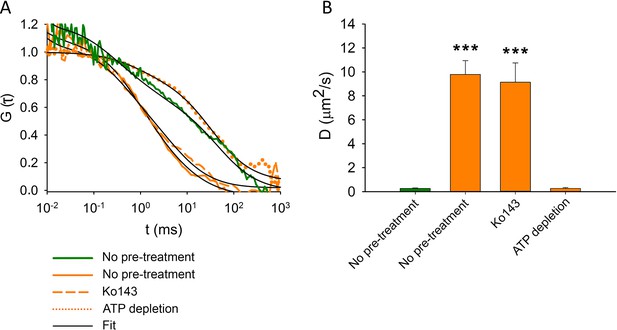
Fluorescence autocorrelation functions (A) and diffusion constants (B) of ABCG2-GFP (green) and MX (orange) in the plasma membrane of intact MDCK cells.
ABCG2-GFP expressing cells without pre-treatments or following ATP depletion or Ko143 pre-treatment were stained with 100 nM MX for 15 min at 37°C. Each bar shows the mean ± SD for n = 50–100 cells from at least three independent measurements. Significant differences compared to the diffusion constant of ABCG2-GFP (green bar) in the plasma membrane of untreated cells are shown by ***: p < 0.001.
-
Figure 7—source data 1
Source Data to Figure 7B.
- https://cdn.elifesciences.org/articles/83976/elife-83976-fig7-data1-v1.xlsx
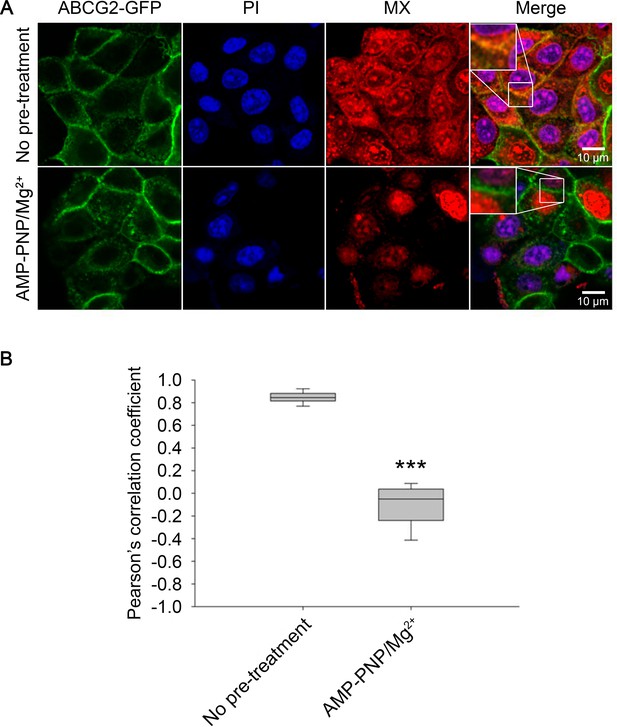
Substrate affinity changes of ABCG2 in permeabilized cells.
SLO-permeabilized ABCG2-GFP expressing cells were pre-treated or not with 5 mM AMP-PNP/Mg2+ for 15 min and then stained with 500 nM MX for 15 min at 37°C. Permeabilized cells were identified by PI staining. Box and whisker plots show Pearson’s correlation coefficients of n > 150 cells from three independent experiments. ***p < 0.001 by Kolmogorov-Smirnov test. MX, mitoxantrone; PI, propidium iodide; SLO, streptolysin-O.
-
Figure 8—source data 1
Source Data to Figure 8B.
- https://cdn.elifesciences.org/articles/83976/elife-83976-fig8-data1-v1.xlsx
Tables
Apparent nucleotide affinities (KA) determined in 5D3-reactivity experiments.
| Treatment | Conditions | KA (mM ± SD) | n | Statistical comparisons to ATP/Mg2+ (1st row) | Figure | |
|---|---|---|---|---|---|---|
| Temp. | Mg2+ | |||||
| ATP | 37°C | + | 4.15 ± 1.08 | 11 | – | Figure 3A |
| ATP+Vi | 37°C | + | 0.34 ± 0.25 | 11 | p < 0.001 | Figure 3A |
| ATP+BeFx | 37°C | + | 0.40 ± 0.06 | 3 | p < 0.001 | Figure 3B |
| ATP | 37°C | − | 3.69 ± 0.49 | 7 | ns | Figure 3C |
| ATP+Vi | 37°C | − | 3.77 ± 0.38 | 4 | ns | Figure 3C |
| ATP+BeFx | 37°C | − | 3.20 ± 0.15 | 3 | ns | Figure 3D |
| ATP | 4°C | + | 4.38 ± 0.24 | 6 | ns | Figure 3E |
| ATP+Vi | 4°C | + | 4.01 ± 0.23 | 3 | ns | Figure 3E |
| ATP+BeFx | 4°C | + | 4.06 ± 0.46 | 3 | ns | Figure 3F |
| AMP-PNP | 37°C | + | 5.29 ± 0.22 | 2 | ns | Figure 3G |
| AMP-PNP+Vi | 37°C | + | 4.91 | 1 | nd | Figure 3G |
| AMP-PNP+BeFx | 37°C | + | 5.88 | 1 | nd | Figure 3H |
| ADP | 37°C | + | 7.37 ± 2.31 | 6 | p < 0.001 | Figure 3I |
| ADP+Vi | 37°C | + | 0.44 ± 0.10 | 3 | p < 0.001 | Figure 3I |
| ADP+BeFx | 37°C | + | 0.57 ± 0.32 | 3 | p < 0.001 | Figure 3J |
-
Table 1—source data 1
Source Data to Table 1.
- https://cdn.elifesciences.org/articles/83976/elife-83976-table1-data1-v1.xlsx
| Reagent type or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | 5D3 (mouse monoclonal) | Hybridoma was donated by Brian P. Sorrentino | Prepared from hybridoma supernatant by affinity chromatography | |
| Antibody | BXP-21 (mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-58222 | (1:2500 dilution) |
| Antibody | C-2 (mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-8432 | (1:2500 dilution) |
| Antibody | Goat anti-mouse IgG-HRP (polyclonal) | Santa Cruz Biotechnology | Cat# sc-2005 | (1:2500 dilution) |
| Cell line (Canis familiaris, dog) | MDCK II, (epithelial-like cells from kidney distal tubule) | Obtained from Prof. Gerrit van Meer | ECACC 00062107 | Mycoplasma-free |
| Cell line (Mus musculus, mouse) | 5D3 hybridoma | Donated by Brian P. Sorrentino | Mycoplasma-free | |
| Chemical compound, drug | Mitoxantrone | Sigma-Aldrich | Cat# M6545 | |
| Chemical compound, drug | Ko143 | Sigma-Aldrich | Cat# K2145 | |
| Chemical compound, drug | Quercetin | Sigma-Aldrich | Cat# Q4951 | |
| Chemical compound, drug | Estrone-3-sulfate | Sigma-Aldrich | Cat# E9145 | |
| Chemical compound, drug | ADP | Sigma-Aldrich | Cat# A2754 | |
| Chemical compound, drug | AMP-PNP | Sigma-Aldrich | Cat# A2647 | |
| Chemical compound, drug | ATP | Sigma-Aldrich | Cat# A2383 | |
| Chemical compound, drug | Sodium-orthovanadate | Sigma-Aldrich | Cat# S6508 | |
| Chemical compound, drug | Protease Inhibitor Cocktail | Sigma-Aldrich | Cat# P2714 | |
| Chemical compound, drug | DL-Dithiothreitol | Sigma-Aldrich | Cat# D0632 | |
| Software, algorithm | Flowing software | Turku Centre for Biotechnology | https://bioscience.fi/services/cell-imaging/flowing-software/ | |
| Software, algorithm | MATLAB | Mathworks Inc. | https://www.mathworks.com/products/matlab.html | |
| Software, algorithm | QuickFit 3.0 | https://biii.eu/quickfit-3 | ||
| Software, algorithm | SigmaPlot | Systat Software Inc. | https://systatsoftware.com/sigmaplot/ | |
| Other | Eight-well chambered coverslip plate | ibidi GmbH | Cat# 80826-90 | Imaging chamber for microscopy |
| Other | Streptolysin-O (SLO) from Streptococcus pyogenes | Sigma-Aldrich | Cat# S5265 | (250 U/ml) |
| Other | Alexa 647 succinimidyl ester | Life Technologies Inc. | Cat# A20006 | Fluorescent dye |
| Other | Propidium iodide | Sigma-Aldrich | Cat# P4170 | Fluorescent dye |
| Other | SuperSignal West Pico PLUS Chemiluminescent Substrate | Thermo Fisher Scientific | Cat# 34579 | Reagent for protein detection in western blot analysis |





