Native functions of short tandem repeats
Figures
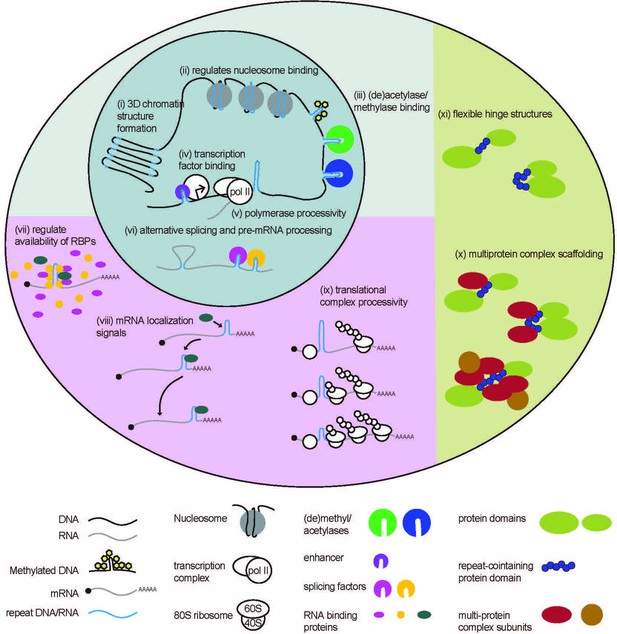
Native functions of genomic repeats.
Repeats in DNA influence larger 3D chromatin structures, regulate binding of nucleosomes and (de)acetylases and (de)methylases. They also influence transcription factor binding and polymerase processivity to affect downstream RNA production. Repeats in RNA can affect pre-mRNA processing such as alternative splicing and can affect RNA binding protein function through direct or indirect sequestration. Repeats in 3’ UTRs serve as localization signals, directing mRNA transport. Repeats in 5’ UTRs regulate translational output by impeding ribosome processivity. Repeating units in proteins can provide structural flexibility within a protein or serve as binding sites for the formation of multi-protein complexes.
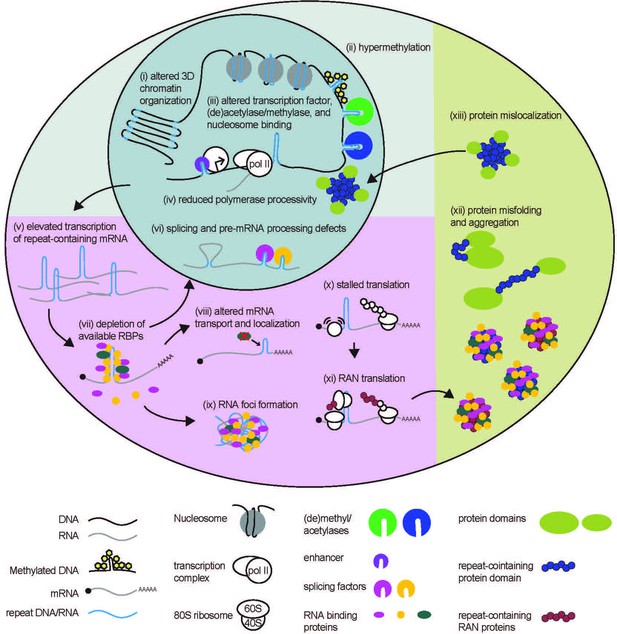
STR-associated toxicity in Repeat Expansion Disorders.
Repeat expansions can alter global 3D chromatin structure, and influence transcription via blocking or enhancing binding of nucleosomes, (de)acetylases, (de)methylases, and transcription factors. Expanded repeats may also impede polymerase processivity. In some cases, elevated transcription of repeat expansion RNA can lead to depletion of RNA-binding proteins. Depletion of these proteins can impact many processes to which they contribute, including pre-mRNA splicing and processing, and mRNA localization. Expanded repeat RNA and bound RBPs can also aggregate into RNA foci, causing toxicity. Expanded repeat RNA can stall translational complexes, leading to repeat-associated non-AUG (RAN) translation, and contribute to the production of polymeric proteins. Polymeric proteins are aggregate prone. Longer polymeric stretches in native proteins may also cause dysfunction by preventing proper protein folding or causing the folded protein to mis-localize within the cell.
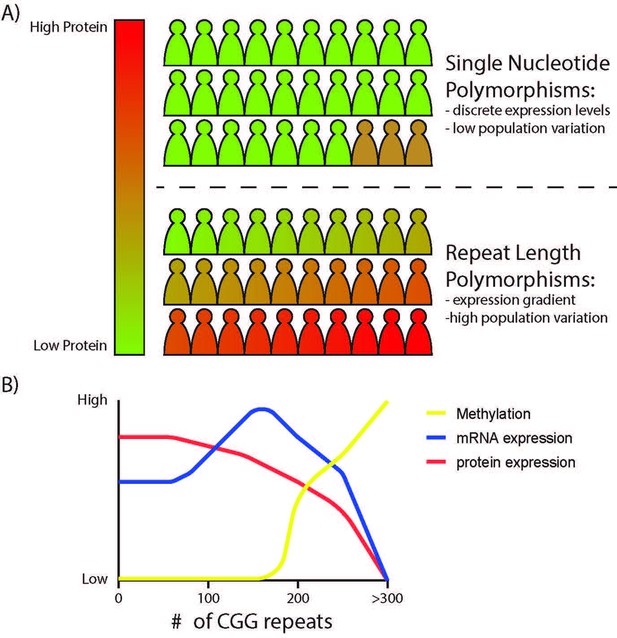
The effects of STR variation on local gene expressivity.
(A) Bi-allelic variation in a gene through single nucleotide polymorphisms often result in small and discrete differences in gene expression, offering limited phenotypic differences across a population with slow evolutionary timescales. In contrast, STRs in promoters and 5’UTRs can influence protein expression over a broader dynamic range, with an inverse correlation between repeat length and protein output within transcribed regions and with differential effects on transcription dependent on the repeat and local epigenetic context. Unstable repeats change rapidly from generation to generation (and even within an individual through somatic variation), creating a mechanism by which mRNA or protein expression can vary broadly and subtly across a population, offering greater genetic and phenotypic diversity and a greater propensity for disease-causing aberrancies at the extremes. (B) Predicted effects of CGG repeat length on FMR1 gene expression. CGG repeat length influences FMR1 promoter epigenetic state (more open chromatin with initial expansion, then DNA methylation and closed chromatin at >200 CGG repeats), FMR1 mRNA expression, and FMRP protein expression across the polymorphic range.





