A human-specific motif facilitates CARD8 inflammasome activation after HIV-1 infection
Figures
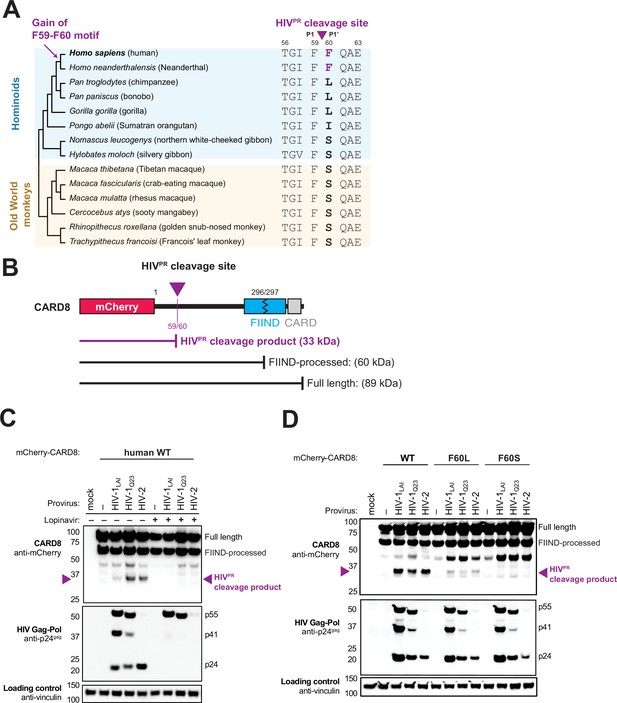
The F59-F60 motif allows human CARD8 to detect protease activity from multiple HIV strs.
(A) Phylogenetic alignment of primate CARD8 protein sequences. The HIV protease (HIVPR) cleavage site is indicated by a purple triangle between F59 (P1) and F60 (P1’). Numbering is based on human CARD8. (B) Depiction of the mCherry-CARD8 used in cleavage assays in (C and D). The predicted molecular weights (kDa) for full-length, FIIND-processed, or HIVPR cleavage products are indicated. FIIND, function-to-find domain; CARD, caspase activation and recruitment domain. (C) HEK293T cells were transfected with a construct encoding N-terminally mCherry-tagged wildtype (WT) CARD8 and indicated HIV proviral constructs, HIV-1LAI, HIV-1Q23, or HIV-2ROD, in the presence (‘+’) or absence (‘–’) of 10 µM lopinavir, an HIVPR inhibitor. Top: Immunoblotting for anti-mCherry to detect the mCherry-CARD8 fusion protein. The full-length and FIIND-processed bands are indicated as well as the HIVPR cleavage product. The band at ~45 kDa is the result of cleavage by the 20S proteasome (Hsiao et al., 2022). Middle: Immunoblotting with an anti-p24gag antibody showing Gag cleavage products p41gag and p24gag, and/or full-length Gag, p55gag. Bottom: Immunoblotting with anti-vinculin as a loading control. (D) HEK293T cells were transfected with a construct encoding N-terminally mCherry-tagged WT, F60L, or F60S CARD8 and indicated HIV proviral constructs. Immunoblotting and labeling of the blots as in (C).
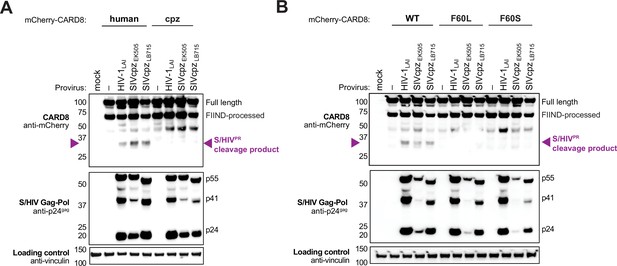
Natural variation in CARD8 alters sensing of SIVcpzPR activity.
(A) HEK293T cells were transfected with a construct encoding N-terminally mCherry-tagged human or chimpanzee (cpz) CARD8 and indicated provirus constructs. Immunoblotting was carried out for CARD8 cleavage, HIV/SIV protease (S/HIVPR) activity, and vinculin (loading control) as indicated. The S/HIVPR cleavage product is indicated by a purple triangle. FIIND, function-to-find domain. (B) HEK293T cells were transfected with a construct encoding N-terminally mCherry-tagged wildtype (WT), F60L, or F60S CARD8 and indicated proviral constructs. Immunoblotting was performed as in (A).
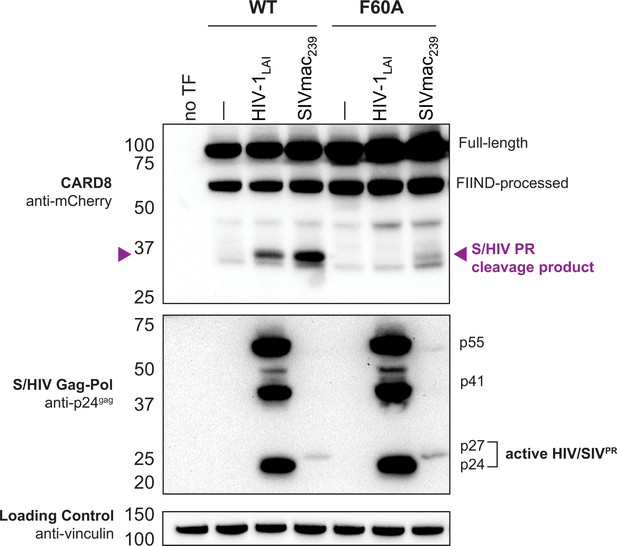
SIVmac cleaves wildtype (WT) human CARD8.
HEK293T cells were transfected with a construct encoding N-terminally mCherry-tagged WT or F60A human CARD8 and indicated provirus constructs. Immunoblotting was carried out for CARD8 cleavage, HIV/SIV protease (S/HIVPR) activity, and vinculin (loading control) as indicated. The anti-p24gag antibody (for S/HIVPR activity) poorly cross-reacts with SIVmac239. The S/HIVPR cleavage product is indicated by a purple triangle. We note that SIVmac239PR still partially cleaves CARD8 F60A. FIIND, function-to-find domain.
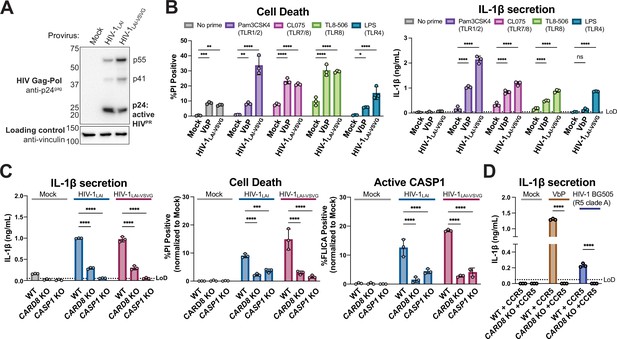
HIV-1 infection activates the CARD8 inflammasome in THP-1 cells.
(A) THP-1 cells were mock infected or infected with HIV-1LAI or HIV-1LAI-VSVG, yielding 8% and 53% p24gag+ cells after 24 hr, respectively. Immunoblotting using cytoplasmic lysates was carried out for HIV protease (HIVPR) activity, and vinculin (loading control) as indicated 24 hr post-infection. (B) THP-1 cells were either left unprimed or primed with different Toll-like receptor (TLR) agonists 4–6 hr before treatment with 10 µM VbP or infection with HIV-1LAI-VSVG at a multiplicity of infection (MOI) such that 30–50% were p24gag+ after 24 hr. Inflammasome responses were measured 24 hr following VbP treatment or HIV-1 infection. Left: Cell death is reported as the percent of propidium iodide (PI) positive cells. Right: Interleukin (IL)-1β levels were measured using the IL-1R reporter assay. (C) Wildtype (WT), CARD8 knockout (KO), or caspase-1 (CASP1) KO THP-1 lines were primed with Pam3CSK4 then challenged with either HIV-1LAI or HIV-1LAI-VSVG at an MOI such that 30–50% of WT cells were p24gag+ after 24 hr. Subsequent inflammasome activation was assayed 24 hr post-infection. Left and middle: Cell death and IL-1β levels were measured as in (A). Right: Active CASP1 was measured with CASP1-specific FLICA dye. (D) WT or CARD8 KO THP-1s overexpressing CCR5 were primed and treated with 10 µM VbP or infected with HIV-1BG505 for 24 hr such that ~30% of cells were p24gag+ then probed for inflammasome activation via IL-1β secretion. HIV-1BG505 is a CCR5 tropic strain in clade A. The dotted line indicates limit of detection (LoD). Datasets represent mean ± SD (n=3 biological replicates). p-Values were determined by two-way ANOVA with Dunnett’s (B–C) or Sidak’s (D) test using GraphPad Prism 9. ns = not significant, *p<0.05,**p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 3—source data 1
Tables of source data for propidium iodide uptake, FLICA, and IL-1β secretion.
- https://cdn.elifesciences.org/articles/84108/elife-84108-fig3-data1-v2.zip

Functional validation of CARD8 knockout (KO) THP-1 cells.
(A) Immunoblot of wildtype (WT) or CARD8 KO THP-1 lines was carried out for CARD8 expression using endogenous antibody against CARD8 C-terminal domain (CTD) and loading control (vinculin). FIIND, function-to-find domain. (B) WT or CARD8 KO THP-1 cells were primed with Pam3CSK4 for 4–6 hr then treated with 10 µM VbP for 24 hr or 5 µg/mL nigericin for 3 hr before probing for interleukin (IL)-1β secretion (left) or propidium iodide (PI) dye uptake (right). The dotted line indicates limit of detection (LoD). Datasets represent mean ± SD (n=3 biological replicates). p-Values were determined by two-way ANOVA Sidak’s test using GraphPad Prism 9. ns = not significant, *p<0.05,**p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 3—figure supplement 1—source data 1
Tables of source data for propidium iodide uptake and IL-1β secretion.
- https://cdn.elifesciences.org/articles/84108/elife-84108-fig3-figsupp1-data1-v2.zip
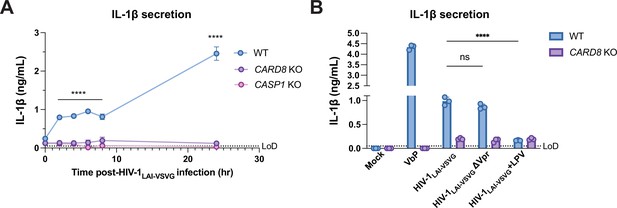
Incoming and outgoing HIVPR ar responsible for CARD8 inflammasome activation.
(A) Wildtype (WT), CARD8 knockout (KO), or caspase-1 (CASP1) KO THP-1 cells were primed overnight with Pam3CSK4 and then infected with HIV-1LAI-VSVG. Supernatant was collected at 0, 2, 4, 6, 8, and 24 hr post-infection to measure interleukin (IL)-1β secretion. Cells were infected at viral concentration such that ~70% of cells were p24gag-positive after 24 hr. (B) WT THP-1 cells primed with Pam3CSK4 were challenged with WT or mutant HIV-1LAI-VSVG or WT virus preincubated in 5 µM lopinavir (LPV) for 30 min prior to infection. HIV-1LAI-VSVG ΔVpr has a frameshift mutation in Vpr. Dotted line indicates limit of detection (LoD). Datasets represent mean ± SD (n=3 biological replicates). p-Values were determined by two-way ANOVA with Dunnett’s test using GraphPad Prism 9. ns = not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 4—source data 1
Tables of source data for IL-1β secretion.
- https://cdn.elifesciences.org/articles/84108/elife-84108-fig4-data1-v2.zip
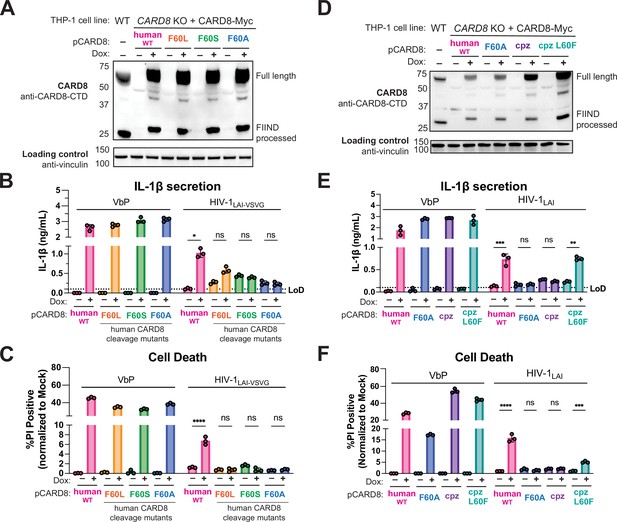
HIV-1 inflammasome activation is dependent on a human-specific motif in CARD8.
(A) CARD8 knockout (KO) THP-1 lines complemented with different doxycycline (dox)-inducible CARD8 variants (pCARD8) were left uninduced or induced for 18 hr. Immunoblot of wildtype (WT) or complemented CARD8 KO THP-1 lines treated with (‘+’) or without (‘–’) dox was carried out for CARD8 expression using endogenous antibody against CARD8 C-terminal domain (CTD) and loading control (vinculin). FIIND, function-to-find domain. (B–C) Complemented CARD8 KO lines were left uninduced or dox-induced as described in (A) and then primed for 4–6 hr with Pam3CSK4 and treated with either 10 µM VbP or HIV-1LAI-VSVG then assessed for (B) interleukin (IL)-1β secretion and (C) cell death, respectively. (D) CARD8 KO THP-1 lines complemented with different CARD8 variants were left uninduced or induced for 18 hr. Immunoblot of wildtype (WT) or complemented CARD8 KO THP-1 lines treated with (‘+’) or without (‘–’) dox as described in (A). (E–F) Complemented CARD8 KO lines were induced and primed as described in (B) then treated with either 10 µM VbP or HIV-1LAI then assessed for (E) IL-1β secretion and (F) cell death, respectively. All HIV infections were done at a multiplicity of infection (MOI) such that 30–50% of cells were p24gag+ after 24 hr. Dotted line indicates limit of detection (LoD). Datasets represent mean ± SD (n=3 biological replicates). p-Values were determined by two-way ANOVA with Tukey’s test using GraphPad Prism 9. ns = not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 5—source data 1
Tables of source data for propidium iodide uptake and IL-1β secretion.
- https://cdn.elifesciences.org/articles/84108/elife-84108-fig5-data1-v2.zip
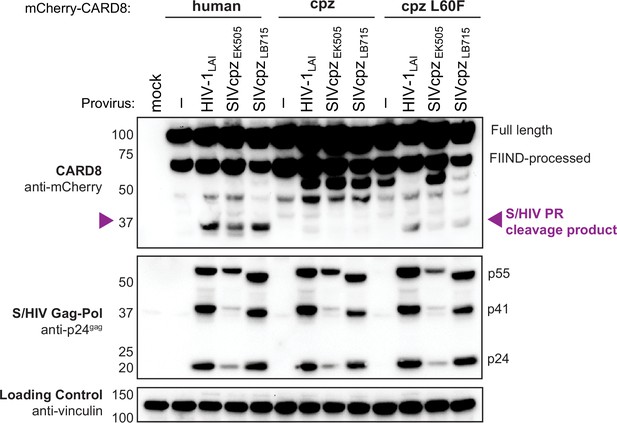
Human FF motif in chimpanzee CARD8 rescues sensing of HIV/SIVPR.
HEK293T cells were transfected with a construct encoding N-terminally mCherry-tagged human, chimpanzee (cpz), or cpz with the human FF motif (L60F) CARD8 and indicated provirus constructs. Immunoblotting was carried out for CARD8 cleavage, HIV/SIV protease (S/HIVPR) activity, and vinculin (loading control) as indicated. The S/HIVPR cleavage product is indicated by a purple triangle. FIIND, function-to-find domain.
Additional files
-
Supplementary file 1
Primers, reagents, and primate CARD8 sequence info.
(a) List of primers, gBlocks, and sgRNA sequences. (b) List of antibodies/reagents. (c) Primate CARD8 gene IDs.
- https://cdn.elifesciences.org/articles/84108/elife-84108-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/84108/elife-84108-mdarchecklist1-v2.pdf
-
Source data 1
- https://cdn.elifesciences.org/articles/84108/elife-84108-data1-v2.zip
-
Source data 2
- https://cdn.elifesciences.org/articles/84108/elife-84108-data2-v2.pdf
-
Source data 3
Uncropped western blots pdf from Figure 2—figure supplement 1, Figure 3—figure supplement 1, Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/84108/elife-84108-data3-v2.pdf





