Recognition of galactose by a scaffold protein recruits a transcriptional activator for the GAL regulon induction in Candida albicans
Figures
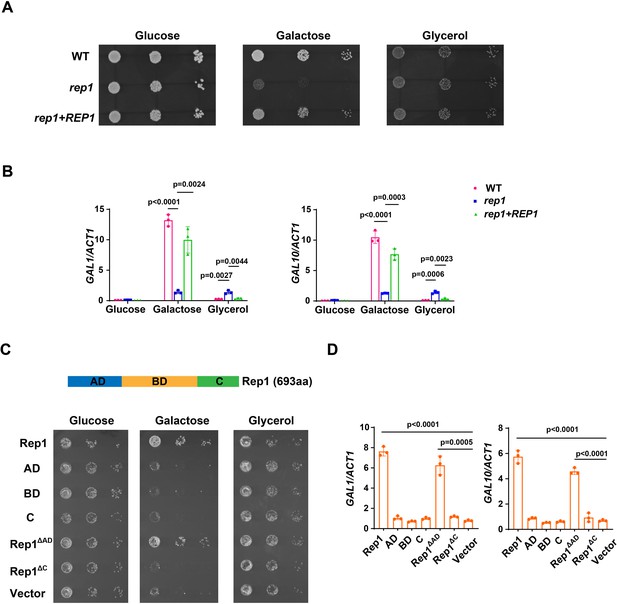
Rep1 is essential for galactose utilization in C. albicans.
(A) Cells of wild type, rep1 mutant or rep1 mutant carrying a wild-type copy of REP1 were diluted 10-fold and spotted onto YNB solid medium containing 25 mM of the indicated sugar. Photographs were taken after 40 hr of growth at 30 °C. (B) qRT-PCR analysis of galactose catabolic genes GAL1 and GAL10 upon galactose induction in the same strains shown in (A). Cells were grown in liquid SC medium with 2% galactose, glucose or glycerol for 2 hr at 30 °C for RNA extraction. (C) Domain structure of Rep1. The transcriptional activation domain (AD) is colored in blue, the DNA binding domain (BD) is colored in orange and the domain located in C-terminus of Rep1 is colored in green. rep1 mutant cells carrying a wild type copy of Rep1, Rep1truncationsAD, BD, C, as well as Rep1ΔC,Rep1ΔAD or vector alone were serially diluted 10-fold and spotted onto YNB solid medium containing 2.5 mM galactose, glucose or glycerol. Graphs were taken after 40 hr of growth at 30 °C. aa: amino acids. (D) The AD domain is unnecessary for Rep1-mediated induction of GAL genes. qRT-PCR analysis of GAL1 and GAL10 upon galactose induction in the same strains shown in (C). (A and C) Representative images of three independent experiments are shown. (B and D) Data shown as means ± SD of three independent experiments. Statistical analysis was performed using an unpaired two-tailed Student’s t-test.
-
Figure 1—source data 1
Raw xlsx files used for analysis of the dataset.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig1-data1-v2.xlsx
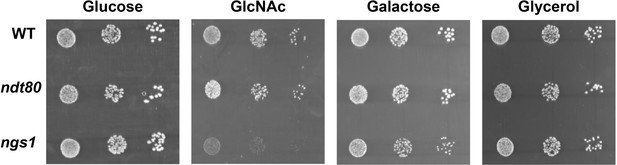
ndt80 or ngs1 mutant did not display a significant defect on galactose utilization in C. albicans.
Cells of wild type, ndt80 or ngs1 mutant were diluted 10-fold and spotted onto YNB solid medium containing 2.5 mM of the indicated sugar. Photographs were taken after 40 hr of growth at 30 °C. Representative images of three independent experiments are shown.
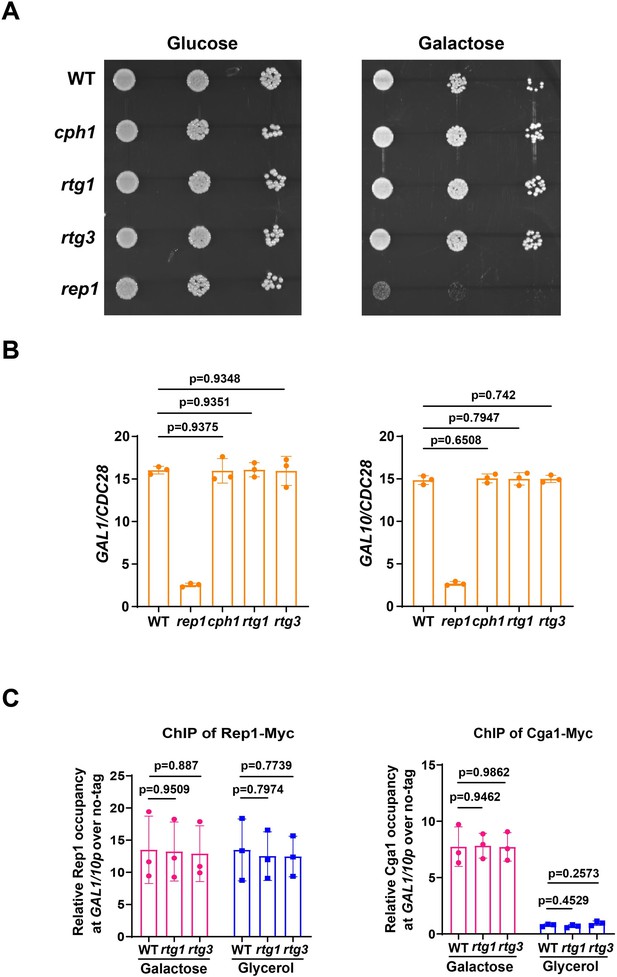
Rtg1 or Rtg3 is not necessary for galactose utilization andRep1-mediated galactose signaling in C. albicans.
(A) Cells of wild type, cph1, rtg1, rtg3, or rep1 mutant were diluted 10-fold and spotted onto YNB solid medium containing 25 mM of the indicated sugar. Photographs were taken after 40 hr of growth at 30 °C. Representative images of three independent experiments are shown. (B) qRT-PCR analysis of galactose catabolic genes GAL1 and GAL10 upon galactose induction in the same strains shown in (A). Cells were grown in liquid SC medium with 2% galactose for 2 hr at 30 °C for RNA extraction. (C) Rtg1 or Rtg3 is not involved in Rep1-Cga1 mode of regulation. ChIP experiments were performed as described in Figure 2A (left) and Figure 3F (right). (B and C) Data shown as means ± SD of three independent experiments.Statistical analysis was performed using an unpaired two-tailed Student’s t-test.
-
Figure 1—figure supplement 2—source data 1
Raw xlsx files used for analysis of the dataset.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig1-figsupp2-data1-v2.xlsx
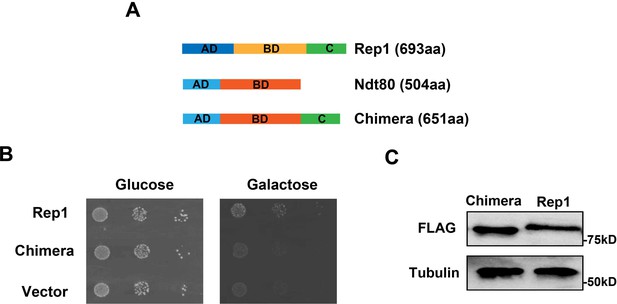
The chimera construct Ndt80-Rep1C fails to rescue the defect of rep1 mutant in galactose signaling.
(A) Schematic depiction of Rep1, Ndt80, and Ndt80-Rep1C chimera construct. aa: amino acids. (B) Introducing the chimera construct into the rep1 mutant could not rescue its growth defect on galactose. Representative images of three independent experiments are shown. (C) Protein level of Chimera construct is comparable to that of Rep1. Representative blots of three independent experiments are shown.
-
Figure 1—figure supplement 3—source data 1
Raw Western blot images with labeled band of interest.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig1-figsupp3-data1-v2.zip
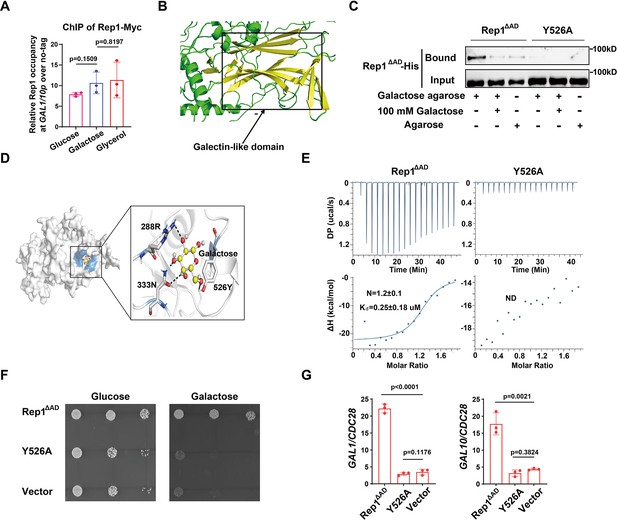
Binding of galactose to Rep1 is required for galactose signaling.
(A) Rep1 is constitutively presented at the GAL1-GAL10 promotor. Overnight culture of wild type cells carrying Rep1-Myc and untagged control was inoculated into SC medium containing 2% indicated sugar at 30 °C for 4 hr for the ChIP experiment. The enrichment is presented as a ratio of GAL1-GAL10 promotor IP (bound/input) over the control region ACT1 IP (bound/input) and is then normalized to the untagged strain. The ChIP data showed the average of three independent experiments with error bars representing the SD. (B) Structure model for Rep1 predicted by AlphaFold2. A galectin-like fold with the characteristic two twisted β-sheets separated by a cleft is shown in Rep1. (C) Rep1ΔAD directly binds to galactose.Recombinant Rep1ΔADand its mutation in Y526 were incubated with Galactose-agarose or control beads at 4 °C overnight in the absence or presence of 100 mM galactose.The input and bound fractions were analysed on Western blots probed with His antibody. Representative blots of three independent experiments are shown. (D) Molecular docking of galactose onto Rep1ΔAD. The highest-ranked docked galactose is shown in the ball-and-stick model, and the protein is shown with a transparent surface. The inset shows how galactose interacts with Rep1ΔAD residues. The galactose and the Rep1ΔAD residues are shown in ball-and-stick and stick models, respectively. Hydrogen bond interactions are shown in dashed lines. (E) ITC binding curves using recombinant Rep1ΔADorits Y526A mutation and galactose (n=3; mean ± SD). (F) Disrupting the binding of Rep1 to galactose leads to a defect in galactose utilization in C. albicans. rep1 mutant cells carrying vector alone, Rep1ΔADor its Y526 mutationwere serially diluted 10-fold and spotted onto YNB solid medium containing 25 mM galactose or glucose. Graphs were taken after 40 hr of growth at 30 °C. Representative images of three independent experiments are shown. (G) qRT-PCR analysis of GAL1 and GAL10 upon galactose induction in the same strains shown in (F). A & G, Data shown as means ± SD of three independent experiments. Statistical analysis was performed using an unpaired two-tailed Student’s t-test.
-
Figure 2—source data 1
Raw xlsx files used for analysis of the dataset.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Raw Western blot images with labeled band of interest.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig2-data2-v2.zip
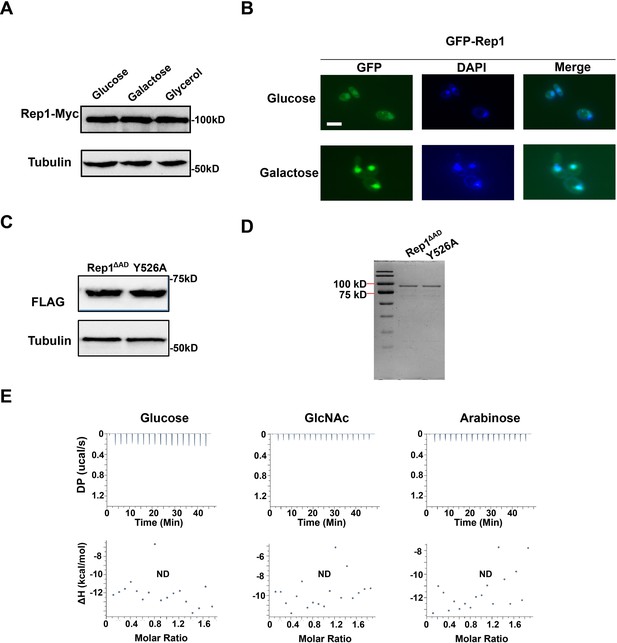
The expression level, DNA-binding property or localization of Rep1 is not regulated by carbon source.
(A) Protein levels of REP1p-Rep1-Myc in SC medium containing 2% glucose, galactose or glycerol were analyzed via western blotting. (B) The rep1 mutant strain carrying GFP-Rep1 under the RHO1 promoter was grown to log phase in liquid SC medium containing 2% galactose or glucose at 30 °C.Rep1 localization was observed by using fluorescence microscopy. Representative images of three independent experiments are shown. Scale bar, 5 μm. (C) The mutation in the putative galactose binding site of Rep1 does not affect its protein level. (D) Coomassie blue staining of purified proteins. (E) ITC binding curves using recombinant Rep1ΔADand glucose, GlcNAc or arabinose (n=3; mean ± SD). (A and C) Representative blots of three independent experiments are shown.
-
Figure 2—figure supplement 1—source data 1
Raw Western blot images and Coomassie stained gel images with the labeled band of interest.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig2-figsupp1-data1-v2.zip
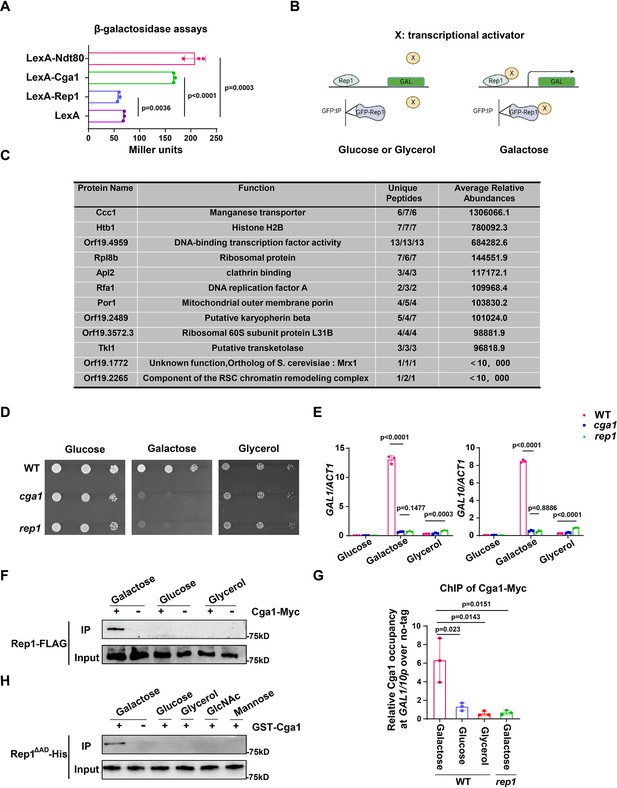
The recognition of galactose by Rep1 enables the recruitment of Cga1 to activate transcription.
(A) Rep1 does not possess the ability to activate transcription, but Cga1 has it. Liquid β-galactosidase assays (in Miller units) of the indicated fusions. (B) A putative model for Rep1-mediated induction of the GAL genes is shown. Protein X represents a transcriptional activator. (C) Putative Rep1 interaction partners specifically in the presence of galactose identified by Mass Spectrometry. Data were obtained from three independent experiments, and the number of unique peptides from each experiment shown together with an average ion score. (D) Cells of indicated strains were diluted 10-fold and spotted onto YNB solid medium containing 25 mM of galactose, glucose or glycerol. Photographs were taken after 40 hr of growth at 30 °C. (E) Cga1 is essential for galactose-inducible expression of GAL genes. Data shown as means ± SD of three independent experiments.(F) Rep1 interacts with Cga1 specifically in galactose in vivo. Overnight culture of wild type cells carrying Rep1-FLAG and Cga1-Myc or Rep1-FLAG alone was diluted 1:50 into SC medium with 2% glucose, galactose or glycerol at 30 °C for 4 hr. Protein lysates were subjected to immunoprecipitation with anti-Myc antibody, and theprecipitated proteins were probed with anti-FLAG antibody. As an inputcontrol, cell lysates were analysed by Western blotting with the anti-FLAGantibody. (G) Rep1 recruits Cga1 to the GAL gene promoter in a galactose-dependent manner. Cells of wild type or rep1 mutant strain carrying Cga1-Myc were diluted at 1:50 into SC medium containing indicated sugars at 30 °C for 4 hr. The enrichment is presented as a ratio of GAL1-GAL10 promotor IP (bound/input) over the control region ACT1 IP (bound/input) and is then normalized to the untagged strain.Data shown as means ± SD of three independent experiments. (H) Rep1 directly interacts with Cga1 in the presence of galactose. Recombinant GST-Cga1 and Rep1∆AD-His were incubated with Glutathione-agarose in the presence of glucose, galactose or glycerol at 4 °C for 2 hr. Samples were assayed by immunoblot with the anti-His antibodies. (A, E and G) Data shown as means ± SD of three independent experiments. Statistical analysis was performed using an unpaired two-tailed Student’s t-test. (F and H) Representative blots or images of three independent experiments are shown.
-
Figure 3—source data 1
Raw xlsx files used for analysis of the dataset.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Raw Western blot images with the labeled band of interest.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig3-data2-v2.zip
-
Figure 3—source data 3
The source file for the table.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig3-data3-v2.zip
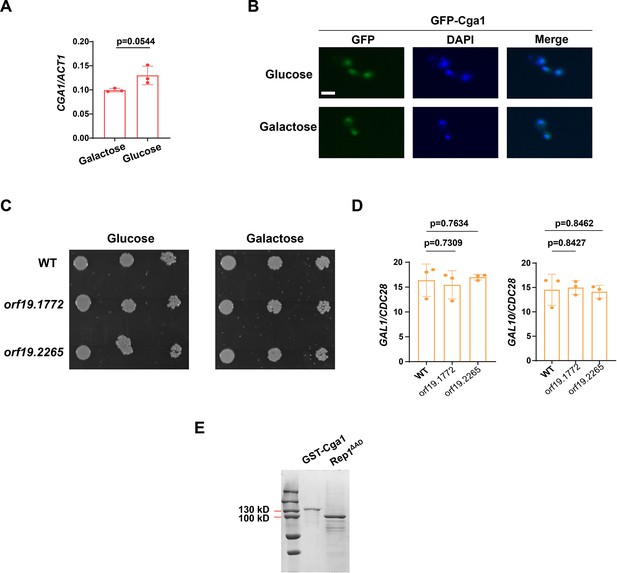
Cga1 is constitutively expressed and accumulated in the nucleus regardless of carbon source.
(A) qRT-PCR analysis of expression levels of CGA1 in SC medium containing 2% glucose or galactose. (B) Localization of GFP-Cga1 in SC medium containing 2% glucose or galactose. Scale bar, 5 μm. (C) Cells of wild type, orf19.1772 or orf19.2265 mutant were diluted 10-fold and spotted onto YNB solid medium containing 25 mM of the indicated sugar. Photographs were taken after 40 hr of growth at 30 °C. Representative images of three independent experiments are shown. (D) qRT-PCR analysis of galactose catabolic genes GAL1 and GAL10 upon galactose induction in the same strains shown in (C). Cells were grown in liquid SC medium with 2% galactose for 2 hr at 30 °C for RNA extraction. (E) Coomassie blue staining of purified proteins.Representative gel images of two independent experiments are shown. (A and D) Data shown as means ± SD of three independent experiments.Statistical analysis was performed using an unpaired two-tailed Student’s t-test.
-
Figure 3—figure supplement 1—source data 1
Raw xlsx files used for analysis of the dataset.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig3-figsupp1-data1-v2.xlsx
-
Figure 3—figure supplement 1—source data 2
Raw Coomassie stained gel images with the labeled band of interest.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig3-figsupp1-data2-v2.zip
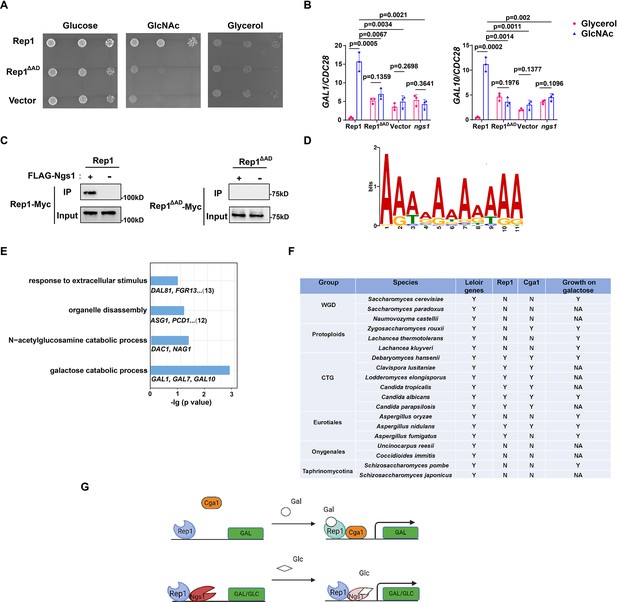
Rep1 provides a scaffold that regulates target genes via its associated partners.
(A) The AD domain of Rep1 is essential for GlcNAc signaling. Cells of the rep1 mutant carrying Rep1,Rep1ΔAD or vector alone were spotted onto YNB solid medium containing 2.5 mM glucose, glycerol or GlcNAc. Photographs were taken after 40 hr of growth at 30 °C. Representative images of three independent experiments are shown. (B) Both the AD domain of Rep1 and Ngs1 are required for the induction of GAL1/GAL10 in GlcNAc. Data shown as means ± SD of three independent experiments.Statistical analysis was performed using an unpaired two-tailed Student’s t-test.(C) The AD domain of Rep1 is required for the recruitment of Ngs1. Protein lysates from wild type cells carrying Rep1-Myc, FLAG-Ngs1 & Rep1-Myc, Rep1ΔAD-Myc, or FLAG-Ngs1 &Rep1ΔAD-Myc were subjected to immunoprecipitation with anti-FLAG antibody, and the precipitated proteins were probed with anti-Myc antibody. As an input control, cell lysates were analysed by Western blotting with the anti-Myc antibody. Representative blots of three independent experiments are shown. (D) The cis-regulatory motif most highly enriched at locations of Rep1 in C. albicans. Motifs were generated using MEME. (E) The major terms after performing GO enrichment analysis. Bar graphs represent the corrected p-value. Representative genes for each GO term are listed below and the number of genes is shown on the right. (F) Presence–absence of Leloir genes, key regulators (Rep1, Cga1) and growth on galactose media. Y, present; N, absent; NA, not available. Growth data were obtained from the review article (Choudhury and Whiteway, 2018). (G) Model of Rep1-mediated transcriptional activation in response to galactose (top) or GlcNAc (bottom). Rep1 recognizes galactose via directly physical interaction, enabling the recruitment of Cga1 to activate transcription. While Rep1 constitutively interacts with GlcNAc sensor Ngs1, the transcriptional activation upon GlcNAc induction is achieved by promoter histone acetylation via GlcNAc binding to Ngs1.
-
Figure 4—source data 1
Raw xlsx files used for analysis of the dataset.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Raw Western blot images with the labeled band of interest.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig4-data2-v2.zip
-
Figure 4—source data 3
The source file for the table.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig4-data3-v2.zip
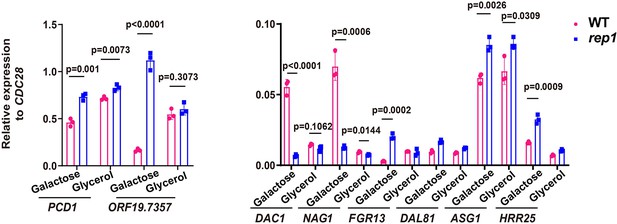
Rep1 binds to its target promoters irrespective of whether those genes are transcriptionally active.
qRT-PCR analysis of expression levels of Rep1 target genes in galactose or glycerol. Data shown as means ± SD of three independent experiments.Statistical analysis was performed using an unpaired two-tailed Student’s t-test.
-
Figure 4—figure supplement 1—source data 1
Raw xlsx files used for analysis of the dataset.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig4-figsupp1-data1-v2.xlsx
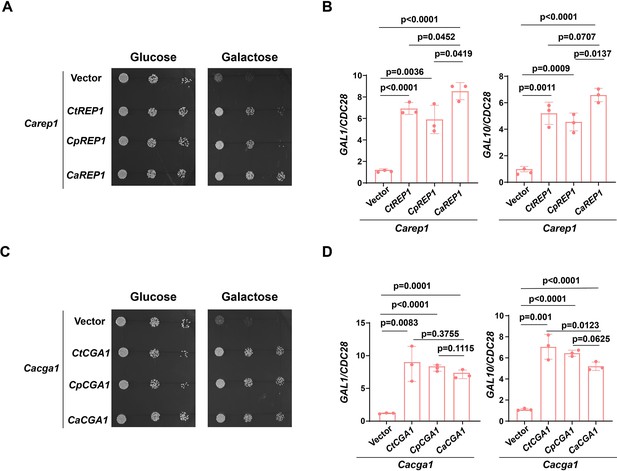
Conservation of Rep1-Cga1 for galactose utilization in CTG species.
(A). Ectopically expressed CtREP1 or CpREP1 suppresses the growth defect of Carep1 mutant on galactose. Dilutions of Carep1 mutant cells carrying CaREP1, CtREP1, CpREP1 or vector alone were grown on YNB solid medium containing 25 mM of the indicated sugar. Photographs were taken after 40 hr of growth at 30 °C. Representative images of three independent experiments are shown. (B). qRT-PCR analysis of galactose catabolic genes GAL1 and GAL10 upon galactose induction in the same strains shown in (A). Cells were grown in liquid SC medium with 2% galactose for 2 h at 30 °C for RNA extraction.Data shown as means ± SD of three independent experiments.Statistical analysis was performed using an unpaired two-tailed Student’s t-test. (C).Spot test experiment of indicated strains was performed as in (A). (D).qRT-PCR experiment of indicated strains was performed as in (B).
-
Figure 4—figure supplement 2—source data 1
Raw xlsx files used for analysis of the dataset.
- https://cdn.elifesciences.org/articles/84155/elife-84155-fig4-figsupp2-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Candida albicans) | REP1 | Candida Genome Database | CR_00,140 W | |
| Gene (Candida tropicalis) | REP1 | Candida Genome Database | CTRG1_01069 | |
| Gene (Candida parapsilosis) | REP1 | Candida Genome Database | CPAR2_800080 | |
| Gene (Candida albicans) | CGA1 | Candida Genome Database | C1_13,320 C | |
| Gene (Candida tropicalis) | CGA1 | Candida Genome Database | CTRG1_03606 | |
| Gene (Candida parapsilosis) | CGA1 | Candida Genome Database | CPAR2_202690 | |
| Strain, strain background (Escherichia coli) | DH5α | Tsingke | TSC-C01 | |
| Strain, strain background (Escherichia coli) | BL21 | Tsingke | TSC-C06 | |
| Antibody | Anti-His (mouse monoclonal) | MBL | D291-3 | 1:5000 |
| Antibody | Anti-c-Myc (rabbit polyclonal) | Sigma-Aldrich | C3956 | 1:5000 |
| Antibody | Anti-FLAG (rabbit polyclonal) | Sigma-Aldrich | F7425 | 1:3000 |
| Antibody | Anti-tubulin (rat monoclonal) | BioRad | MCA78G | 1:3000 |
| Antibody | Anti-mouse IgG, (HRP-linked, mouse polyclonal) | BioRad | 1706516 | 1:3000 |
| Antibody | Anti-rat IgG (HRP-linked, rat polyclonal) | CST | 7077 | 1:5000 |
| Antibody | Anti-rabbit IgG, (HRP-linked, rabbit polyclonal) | Sigma-Aldrich | A3687 | 1:5000 |
| Sequence-based reagent | pBA1-F | This paper | PCR primers | CAACAACAAATACAAAAACAAAGATCT |
| Sequence-based reagent | pBA1-R | This paper | PCR primers | ATACGACTCACATAGGGCGAATTGGGTACC |
| Sequence-based reagent | pET28a-F | This paper | PCR primers | CTCACAGAGAACAGATTGGTGGATCC |
| Sequence-based reagent | pET28a-R | This paper | PCR primers | TCAGTGGTGGTGGTGGTGGTGCTCGA |
| Peptide, recombinant protein | Rep1ΔAD | This paper | purified from E. coli BL21 | |
| Peptide, recombinant protein | Rep1ΔAD Y526A | This paper | purified from E. coli BL21 | |
| Peptide, recombinant protein | GST-Cga1 | This paper | purified from E. coli BL21 | |
| Commercial assay or kit | RNeasy Minikit | Qiagen | 74204 | |
| Commercial assay or kit | GFP-Trap | Chromotek | Gta-20 | |
| Chemical compound, drug | D-Galactose | Sigma-Aldrich | G5388 | |
| Chemical compound, drug | N-Acetyl-D-glucosamine | MP | 1727589 | |
| Chemical compound, drug | D-Glucose | Sigma-Aldrich | G7021 | |
| Chemical compound, drug | Glycerol | Sigma-Aldrich | G5516 | |
| Chemical compound, drug | D-Mannose | Sigma-Aldrich | M2069 | |
| Chemical compound, drug | D-Arabinose | Aladdin | 10323-20-3 | |
| Software, algorithm | MicroCal PEAQ-ITC | Malvern Panalytical | MicroCal ORIGIN | |
| Software, algorithm | AutoDock Vina | AutoDock Vina | ||
| Software, algorithm | Grahpad prism | Grahpad prism | 8.0.3 | |
| Other | DAPI | Solarbio | C0060 | 1 mg/ml |
Additional files
-
Supplementary file 1
(A) Candida albicans strains used in this study. (B) Plasmids used in this study. (C) Primers used in this study.
- https://cdn.elifesciences.org/articles/84155/elife-84155-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/84155/elife-84155-mdarchecklist1-v2.pdf





