A human mitofusin 2 mutation can cause mitophagic cardiomyopathy
Figures
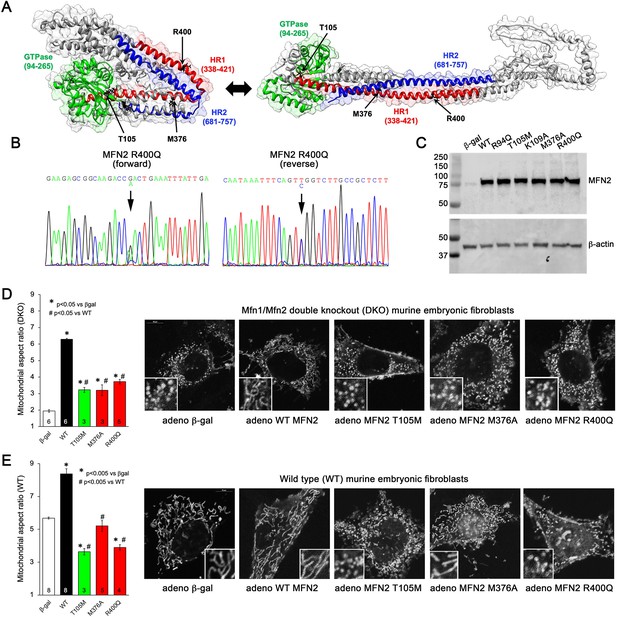
Morphological effects of disease-related MFN2 mutants on mitofusin null and normal mitochondria.
(A) Hypothetical structures of human MFN2 in the basal/closed (left) and active/open (right) conformations. Domains are color-coded: green, GTPase; red, first heptad repeat (HR1); blue, second heptad repeat (HR2). Arrows indicate positions of human disease-linked MFN2 mutants of primary interest. (B) Sequencing pherogram from a hypertrophic cardiomyopathy subject showing heterozygous MFN2 R400Q mutation (arrow). (C) Immunoblot of wild-type (WT) and mutant MFN2s expressed in Mfn1/Mfn2 double knockout (DKO) murine embryonic fibroblasts (MEFs). In all studies, adenoviral β-gal is negative control; β-actin is loading control. (D) Intrinsic fusogenic activity of WT and mutant MFN2 expressed in MFN DKO cells, assayed as increase in mitochondrial aspect ratio compared to β-gal. (E) Same as (D) except MFNs were expressed in WT MEFs to reveal effects on endogenous MFN1 and MFN2. Unless otherwise stated, in all graphs black is WT MFN2, green is GTPase domain MFN2 mutants, red is HR1 domain MFN2 mutants. Numbers in bars are independent experiments for 6–30 cells per experiment. p-Values used ANOVA and Tukey’s test.
-
Figure 1—source data 1
gnomAD MFN2 coding variants.
Red font indicates variants found in CM.
- https://cdn.elifesciences.org/articles/84235/elife-84235-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Mitofusin 2 (75 kDa) and β-actin (45 kDa) expression level in MEFs Wt.
- https://cdn.elifesciences.org/articles/84235/elife-84235-fig1-data2-v1.zip
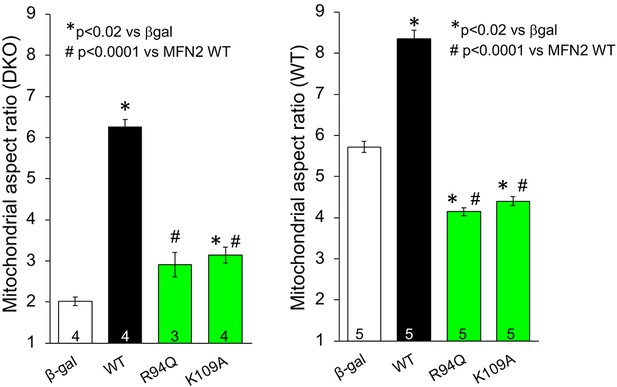
Fusogenic activities of GTPase-defective, CMT2A-linked MFN2 R94Q, and laboratory-engineered K109A in Mfn1/Mfn2 double knockout (DKO) murine embryonic fibroblasts (MEFs) (left) and WT MEFs (right).
β-gal and WT MFN2 are from Figure 1. p-Values calculated using ANOVA and Tukey’s test.
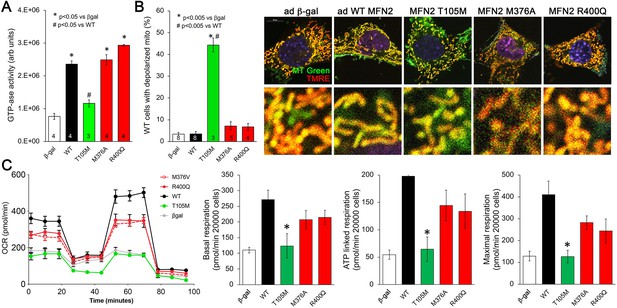
Effects of disease-related MFN2 mutants on catalytic and respiratory function of mitofusin null and normal mitochondria.
(A) GTPase activity of human disease-linked MFN2 mutants expressed in mitofusin double knockout (DKO) murine embryonic fibroblasts (MEFs). (B) Loss of inner membrane polarization in WT MEF mitochondria induced by disease-related MFN2 mutants. To the right are representative MitoTracker Green/TMRE co-stained confocal images. (C) Seahorse studies of oxygen consumption rate (OCR) in MFN DKO MEFs expressing disease-linked MFN2 mutants. (Left) Average data from three experiments per condition. (Right) Group quantitative results for basal, ATP-linked and maximal respiration (pmol/min/20,000 cells). *p<0.05 vs. WT.p-Values for all studies used ANOVA and Tukey’s test.
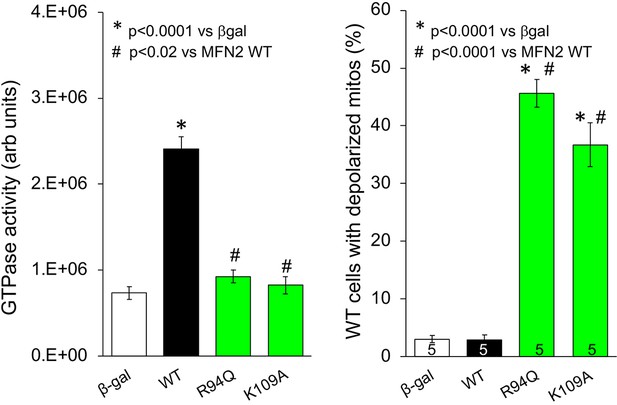
GTPase activity of, and inner membrane depolarization induced by, GTPase domain CMT2A-linked MFN2 R94Q and laboratory-engineered K109A expressed in Mfn1/Mfn2 double knockout (DKO) murine embryonic fibroblasts (MEFs) (left) and WT MEFs (right), respectively.
β-gal and WT MFN2 are from Figure 2. p-Values calculated using ANOVA and Tukey’s test.
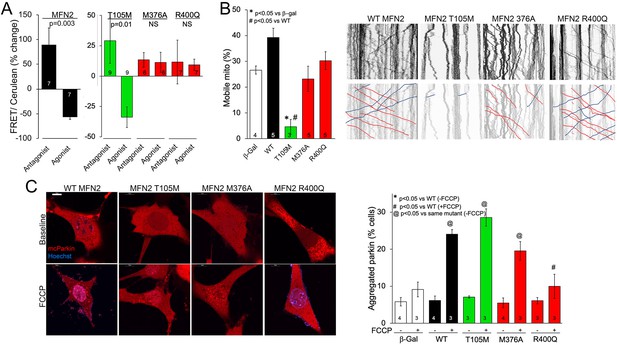
MFN2 R400Q impairs conformational shifting and suppresses mitochondrial Parkin recruitment, but not mitochondrial motility in neuronal axons.
(A) Forster resonance energy transfer (FRET) studies of conformational switching in disease-linked MFN2 mutants expressed in MFN double knockout (DKO) murine embryonic fibroblasts (MEFs). Antagonist peptide normally induces closed conformation (increase in FRET) and agonist peptide open conformation (decrease in FRET). (B) Mitochondrial motility in cultured mouse dorsal root ganglion neuronal processes: representative kymographs are to the right (raw data above; motile mitochondrial emphasized below [red, antegrade; blue, retrograde]).Scale bar 10 μm, Data are n~3 5 DRGs for each independent experiment, statistic is one-way ANOVA. (C) mcParkin (red) recruitment to mitochondria in MFN DKO MEFs expressing MFN2 mutants before (baseline) and after FCCP (left, representative confocal micrographs; right, group quantitative data). p-Values calculated using ANOVA and Tukey’s test.
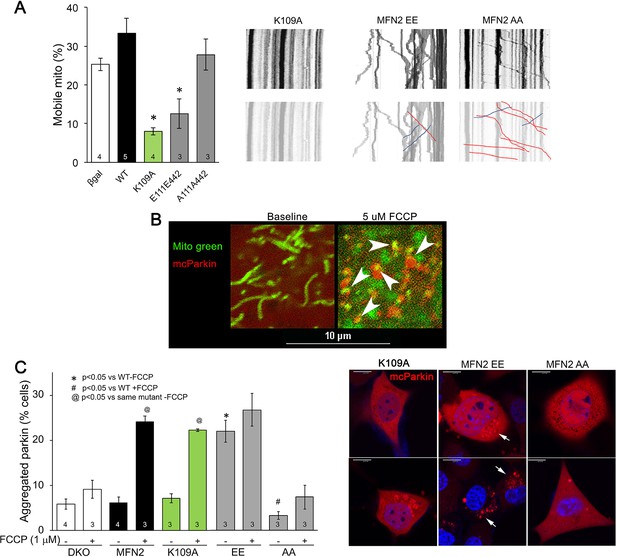
Effects of engineered synthetic MFN2 mutants on mitochondrial Parkin recruitment and mitochondrial motility in neuronal axons.
(A) Mitochondrial motility in cultured dorsal root ganglion (DRG) neuronal processes. Group quantitative data are to the left; representative kymographs are to the right. β-gal and WT MFN2 are from Figure 3. p-Values calculated using ANOVA and Tukey’s test. (B) Representative confocal micrograph showing Parkin localization (red) to mitochondria (green) after FCCP treatment. Arrows point to some co-localization events (yellow). Scale bar 10 μm, Data are n~3 5 DRGs for each independent experiment. (C) Parkin aggregation at mitochondria in MFN2 double knockout (DKO) murine embryonic fibroblasts (MEFs) in the absence (-) and presence (+) of FCCP; representative images to the right. MFN2 K109A is GTPase-defective; MFN2 EE constitutively binds Parkin; MFN2 AA cannot bind Parkin.
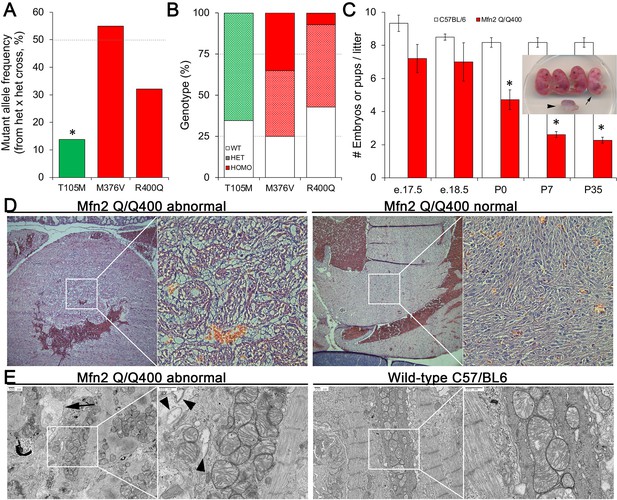
Perinatal cardiomyopathy in Mfn2 Q/Q400 mice.
(A, B) Mutant allele frequencies (A) and genotypes (B) from heterozygous crosses of Mfn2 T105M, M376V, and R400Q knock-in mice (CRISPR/Cas9 knock-in strategies are given in the figure supplement). Expected values are shown as dotted lines. Homozygous mice were absent for T105M and fewer than expected for R400Q. (C) Fetal and early postnatal lethality of homozygous R400Q knock-in mice (Q/Q400) reported as mice/litter at different time points. Controls (white) are C57/Bl6. Inset shows an E18.5 Q/Q400 litter having a degenerated fetus (arrowhead) and a live, non-viable fetus (arrow). (D) Hematoxylin and eosin-stained sections of left ventricular myocardium from P0 Q/Q400 mice with (left) and without (right) cardiomyocyte vacuolization. (E) Ultrastructural studies of Q/Q400 myocardium revealing myofibrillar degeneration (arrow), empty ‘ghost’ mitochondria (arrowheads) and mitochondrial fragmentation; wild-type control is shown for comparison on the right. *p<0.05 vs. expected (A, Fisher’s exact test) or control (C, Student’s t-test).
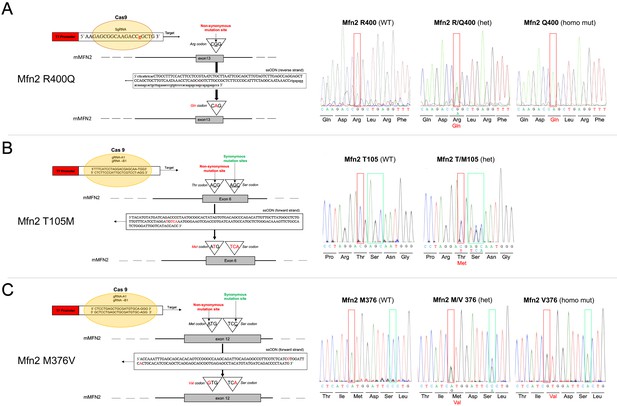
CRISPR engineering of disease-linked human MFN2 mutants into the mouse genome.
(A) Mfn2 R400Q; (B) Mfn2 T105M; and (C) Mfn2 M376V. For each mouse line, the left of the panel depicts the gene editing strategy, and to the right Sanger sequence of the resulting nucleotide changes. Note that no homozygous mice or embryos as early as E12.5 were identified for Mfn2 T105M in >40 breedings.
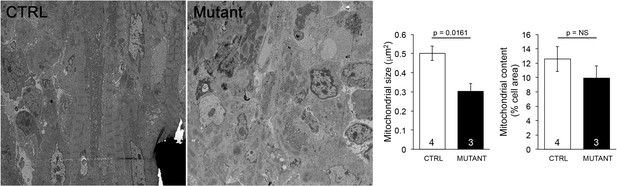
Quantitative mean data for transmission electron micrograph (TEM) studies of Mfn2 Q/Q400 mouse pups.
On the left are representative TEMs of myocardium. To the right are mean ± SEM data. Numbers are individual mice with >10 images per mouse; p-values by unpaired t-test.
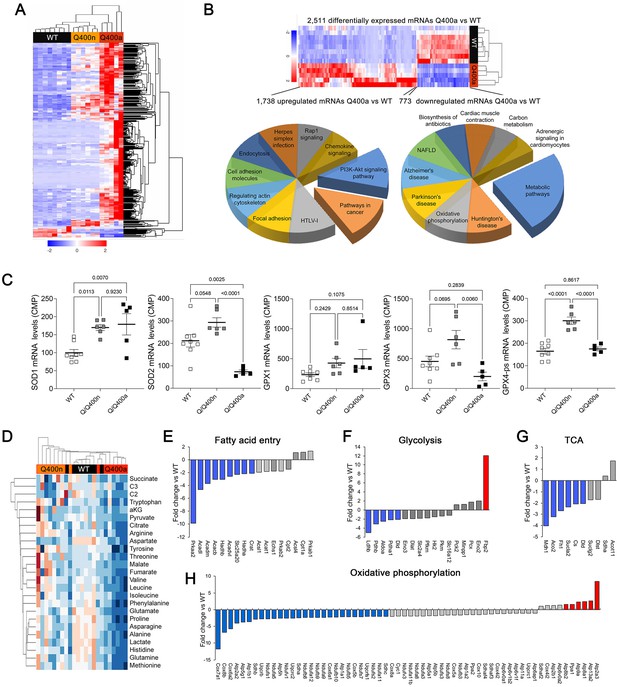
Transcriptional and metabolomic profiling of Mfn2 Q/Q400 mouse hearts identifies metabolic abnormalities characteristic of mitophagy defects.
(A) Heat map of gene expression in individual Q400 and wild-type (WT) mouse hearts. (B) Heat map of anomalous Q400a mouse heart gene expression (top) and pie charts describing major KEGG functionally annotated pathway categories of up- (left) and downregulated (right) transcripts. (C) Individual heart mRNA levels for reactive oxygen species (ROS)-modulating enzymes. SOD, superoxide dismutase; GPX, glutathione peroxidase. Results shown are for the cardiac-expressed isoforms defined as WT CPM >100. Each point is an individual mouse heart. p-Values by ANOVA. (D) Metabolomics heat map showing unsupervised clustering of individual Q/Q400 and WT mouse hearts. (E–H) Relative expression of genes from indicated metabolic pathways. Blue is significantly decreased in MFN2 Q/Q 400a hearts; red is significantly increased; gray is no significant difference.
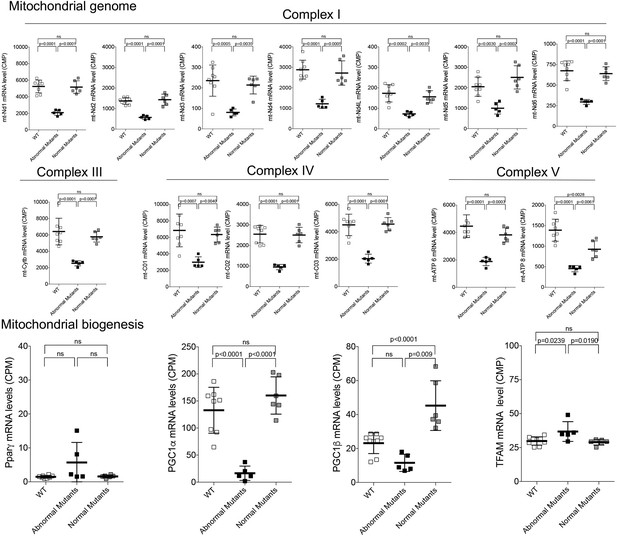
Expression of mitochondrial DNA-encoded respiratory genes (top) and mitochondrial biogenesis genes (bottom) in late fetal Mfn2 Q/Q400 mouse hearts.
Each point is an individual mouse heart. p-Values by ANOVA.
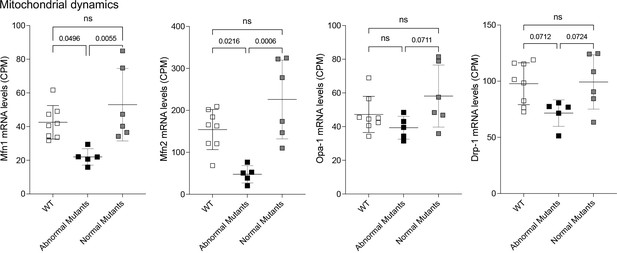
Expression of mitochondrial dynamics factor mRNAs in late fetal Mfn2 Q/Q400 mouse hearts.
Each point is an individual mouse heart. p-Values by ANOVA.
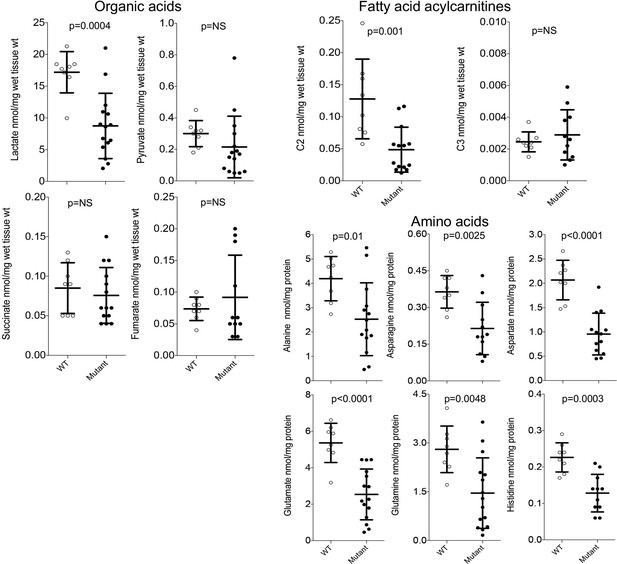
Metabolite levels in Mfn2 Q/Q400 mouse pup hearts.
Each point is an individual mouse heart. ‘Mutant’ indicates both abnormal and normal MFN2 Q400 mutants. p-Values from unpaired t-test.
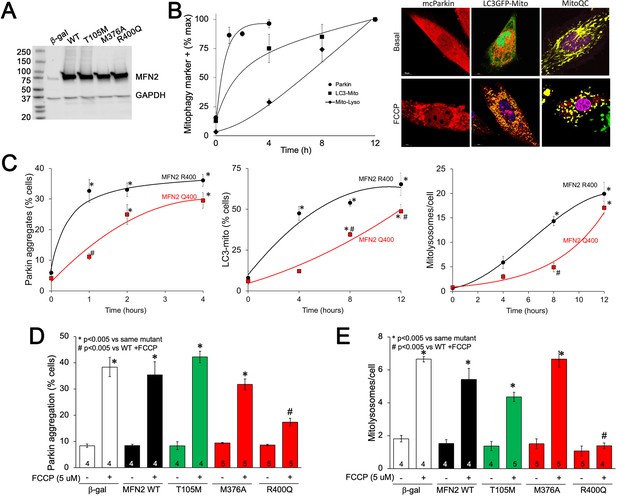
MFN2 Q400 uniquely suppresses early and late mitophagy in cardiomyoblasts.
(A) Adenoviral expression of MFN2s in H9c2 cells. (B) Time courses of mitochondrial Parkin aggregation (circles), mitochondrial engulfment by autophagosomes (squares), and mitochondrial delivery to lysosomes (diamonds) in WT MFN2-expressing H9c2 cells after FCCP stimulation. Representative images are to the right. (C) Comparative time courses for mitochondrial Parkin aggregation (left), mitochondrial engulfment by autophagosomes (middle), and mitochondrial delivery to lysosomes (right) in MFN2 WT (black) and Q400 (red) expressing H9c2 cells. n = 3–4, *p<0.05 vs. pre-FCCP (time 0); #p<0.05 vs. WT at same time point (two-way ANOVA). (D, E) mcParkin aggregation 1 hr after FCCP treatment (D) and mitolysosome formation 8 hr after FCCP treatment (E) of H9c2 cells transduced with MFNs. Experimental n is shown in bars; stats by two-way ANOVA. Representative confocal images are shown in Figure 6—figure supplement 1.
-
Figure 6—source data 1
Mitofusin 2 (75kDa) and GAPDH (36 kDa) expression level in H9c2 cells.
- https://cdn.elifesciences.org/articles/84235/elife-84235-fig6-data1-v1.zip
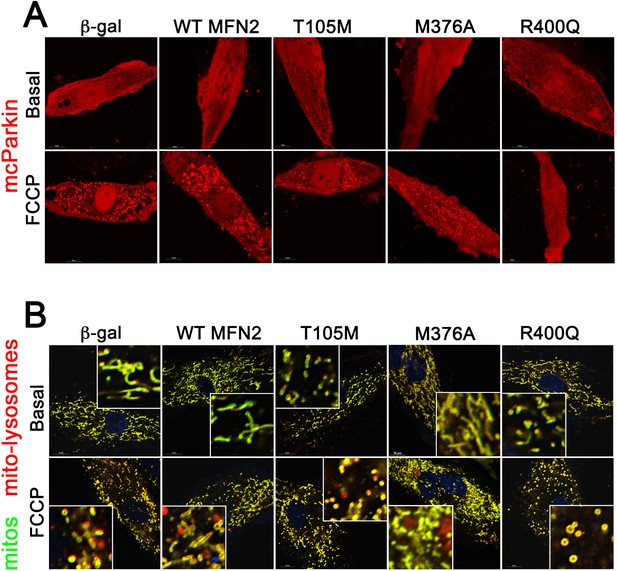
Representative mcParkin (A) and MitoQC (B) confocal images showing mitophagy provoked by FCCP in H9c2 cardiomyoblasts expressing WT and disease-linked MFN2 mutants.
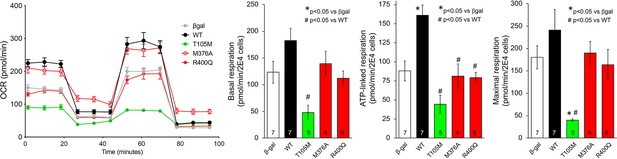
Seahorse studies of oxygen consumption rate (OCR) in cultured H9c2 cardiomyoblasts expressing disease-linked MFN2 mutants.
(Lleft) Average data from 5 to 7 experiments per condition. (Right) Group quantitative results for basal, ATP-linked, and maximal respiration (pmol/min/20,000 cells). Statistical comparisons used ANOVA.
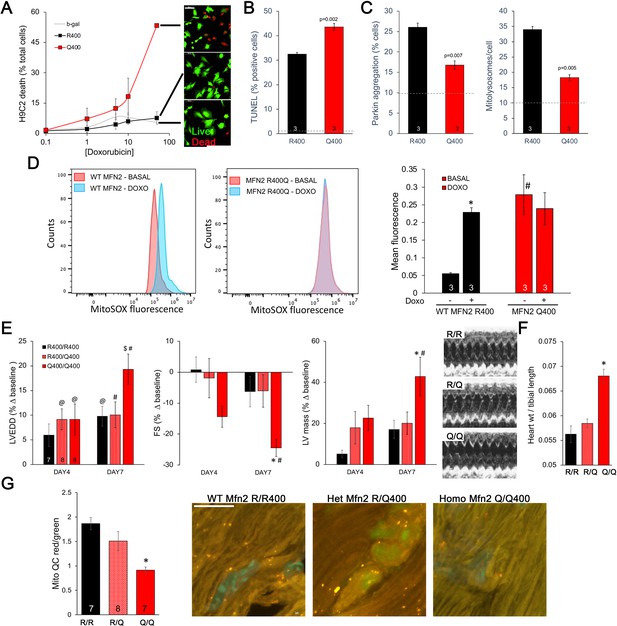
Mfn2 R400Q impairs reactive mitophagy and amplifies cardiomyocyte toxicity induced by doxorubicin in vitro and in vivo.
(A) Cell death curves provoked by increasing concentrations of doxorubicin (overnight treatment) in WT MFN2 (R400) and MFN2 Q400 transduced H9c2 cardiomyoblasts. Representative live-dead images corresponding to each group are to the right; scale bar 100 um. (B) TUNEL labeling of apoptotic H9c2 cardiomyoblasts treated with 10 uM doxorubicin overnight. Dotted line is basal level in absence of doxorubicin. (C) Reactive mitophagy measured as Parkin aggregation (left) and mitolysosome formation (right) in doxorubicin-treated H9c2 cardiomyoblasts. Dotted lines show basal levels in absence of doxorubicin. (D) Flow cytometric analysis of doxorubicin effects on mitochondrial-derived ROS in H9c2 cardiomyoblasts expressing WT MFN2 R400 or mutant MFN2 Q400. Representative (of three independent experiments) histograms are on the left; mean group data are on the right. *p<0.05 vs. same group without doxorubicin; #p<0.05 vs. WT cells treated identically (ANOVA). (E) Echocardiography of WT (R/R400), heterozygous (R/Q400), and homozygous (Q/Q400) knock-in mice 4 and 7 d after receiving doxorubicin (20 ug/g, intraperitoneal injection). LVEDD is left ventricular (LV) end diastolic dimension, which increases with dilated cardiomyopathy; FS is LV fractional shortening, which decreases in dilated cardiomyopathy; calculated LV mass increases in dilated cardiomyopathy. Representative m-mode echos are to the right. (F) Gravimetric heart weights 7 d after doxorubicin, indexed to tibial length. (G) MitoQC visualization of LV cardiomyocyte mitolysosomes. Group mean quantitative data on the left; representative confocal images on the right. Mitolysosomes are bright orange spots; scale bar is 10 um. (E–G): *p<0.05 vs. WT at the same time; $p=0.05–0.075 vs. WT at the same time; #p<0.05 vs. same genotype pre-doxorubicin; @p=0.05–0.075 vs. same genotype pre-doxorubicin; statistical comparisons used one- (F, G) or two-way (E) ANOVA.
Tables
Results of genetic screening for MFN2 mutations in adult cardiomyopathy.
HCM, hypertrophic cardiomyopathy; DCM, dilated cardiomyopathy; MAF, minor allele frequency. MFN2 protein domains correspond to colored regions in Figure 1A.
| MFN2 variant | MFN2 domain | Polyphen 2 | ClinVar | HCM n = 286 | DCM1 n = 398 | DCM2 n = 281 probands (424 cases) | CM MAF n = 965 | gnomAD MAF n = 141,456 | MAF CM vs. gnomAD (p-value) | MAF CM vs. gnomAD (X2 stat) |
|---|---|---|---|---|---|---|---|---|---|---|
| S180R | GTPase | Benign | CMT2A | 1 het | Singleton | 1.591 E-05 | ||||
| V181M | GTPase | Prob dam | CMT2A | 1 family; 5 indiv | Singleton | 3.976 E-06 | ||||
| S263Y | GTPase | Pos dam | 1 het | Singleton | - | - | - | |||
| R274Q | GTPase | Benign | CMT2A | 1 family; 4 indiv | Singleton | 2.486 E-05 | ||||
| G298R | GTPase | Benign | CMT2A | 1 het | Singleton | 2.147 E-03 | ||||
| M393I | HR1 | Benign | Likely benign | 1 het | Singleton | 1.591 E-05 | ||||
| R400Q | HR1 | Prob dam | Uncertain significance | 2 hets | 1 het | 1.55 E-03 | 7.423 E-05 | <0.00001 | 89.595 | |
| Q413P | HR1 | Benign | 1 family; 1 indiv | Singleton | 1.193 E-05 | |||||
| R468H | inter | Pos dam | CMT2A | 1 het | 2 families; 5 indiv | 1.55 E-03 | 2.177 E-03 | NS | 0.3431 | |
| V705I | HR2 | Benign | Likely benign | 2 hets | 6 families; 15 indiv | 4.14 E-03 | 6.865 E-03 | NS | 2.0848 | |
| A716T | HR2 | Pos dam | CMT2A | 1 family; 1 indiv | Singleton | 1.308 E-04 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Mfn-2 | NCBI Gene | Gene ID: 170731 | MFN2 ENSMUSG00000029020 |
| Gene (human) | MFN-2 | NCBI Gene | Gene ID: 9927 | MFN2 ENSG00000116688 |
| Genetic reagent (M. musculus) | Mfn2 R400Q knock-in mice | DornLab | Knock-in mice generated with point mutation in position 400 | |
| Genetic reagent (M. musculus) | Mfn2 T105M knock-in mice | DornLab | Knock-in mice generated with point mutation in position 105 | |
| Genetic reagent (M. musculus) | Mfn2 M376V knock-in mice | DornLab | Knock-in mice generated with point mutation in position 376 | |
| Genetic reagent (M. musculus) | C57BL/6J mice | C57Bl/6 | The Jackson Laboratory: 000664 | C57Bl/6 |
| Cell line H9c2 (rat myoblast) | Cardiomyoblast | ATCC | CRL-1446 | Rat embryonic cardiomyoblast |
| Cell line Mfn1/Mfn2 null (M. musculus) | Mfn1 and Mfn2 double knock out MEFs | ATCC | CRL-2993 | Murine embryonic fibroblasts |
| Cell line DRG neuronal cells | Adult mouse dorsal root ganglion | From 8- to 12-week-old C57BL/6J using enzymatic dissociation. | Franco et al., 2020 | |
| Cell line mouse embryonic fibroblast | MEFs WT | From E.14.5 pups from C57BL/6J using enzymatic dissociation | https://pubmed.ncbi.nlm.nih.gov/18265203/ | |
| Transfected construct (human adenovirus type 5 dE1/E3) | Adenovirus Mito-Ds-Red2 | Signagen | Cat# 12259 | |
| Transfected construct (human adenovirus type 5 dE1/E3) | Adenovirus Cre-recombinase | Vector Biolabs | Cat# 1794 | |
| Transfected construct (human adenovirus type5 dE1/E3) | Ad-MFN2T105M | DornLab | Franco et al., 2022 | |
| Transfected construct (human adenovirus type 5 dE1/E3) | Ad-MFN2R94Q | DornLab | Franco et al., 2022 | |
| Transfected construct (human adenovirus type 5 dE1/E3) | Ad-MFN2R400Q | DornLab | Human adenovirus type 5 (dE1/E3), with Mfn2R400Q | |
| Transfected construct (human adenovirus type 5 dE1/E3) | Ad-MFN2K109A | DornLab | Rocha et al., 2018 | |
| Transfected construct (human adenovirus type 5 dE1/E3) | Ad-MFN2M376A | DornLab | Rocha et al., 2018 | |
| Transfected construct (human adenovirus type 5 dE1/E3) | Ad-MFN2WT | DornLab | Franco et al., 2022 | |
| Transfected construct (human adenovirus type 5 dE1/E3) | Ad-hFRETMFN22WT | DornLab | Rocha et al., 2018 | |
| Transfected construct (human adenovirus type 5 dE1/E3) | Ad-hFRETMFN2R94Q | DornLab | Franco et al., 2022 | |
| Transfected construct (human adenovirus type 5 dE1/E3) | Ad-hFRETMFN2K109A | DornLab | Human adenovirus type 5 (dE1/E3), with Mfn2K109A | |
| Transfected construct (human adenovirus type 5 dE1/E3) | Ad-hFRETMFN2M376A | DornLab | Franco et al., 2022 | |
| Transfected construct (human adenovirus type 5 dE1/E3) | Ad-hFRETMFN2T105M | DornLab | Franco et al., 2022 | |
| Transfected construct (human adenovirus type 5 dE1/E3) | Ad-LC3-GFP | DornLab | Human adenovirus type 5 (dE1/E3), with LC3 for autophagy | |
| Transfected construct (human adenovirus type 5 dE1/E3) | Ad-mCherry-Parkin | Gift from Dr. Åsa Gustafsson | N/A | |
| Transfected construct (human adenovirus type 5 dE1/E3) | Ad-mitoQC | DornLab | Human adenovirus type 5 (dE1/E3), with mitoQC for mitophagy | |
| Antibody | Anti-Mfn-2 (mouse monoclonal) | Abcam | Cat# ab56889 | 1:1000 |
| Antibody | Anti-COX-IV (rabbit polyclonal) | Abcam | Cat# ab16056 | 1:1000 |
| Antibody | Anti-GAPDH (mouse monoclonal) | Abcam | Cat# ab82245 | 1:3000 |
| Antibody | Anti-β-actin (unconjugated monoclonal) | Proteintech | Cat# 66009-1 | 1:3000 |
| Antibody | Goat anti-rabbit IgG | Thermo Fisher | Cat# 31460 | 1:3000 |
| Antibody | Peroxidase-conjugated anti-mouse IgG | Cell Signaling | Cat# 7076S | 1:3000 |
| Sequence-based reagent | mMFN2R400Q knock-in mouse (forward) | DornLab | Knock-in mice generated with point mutation in position 400 | GGCATGTATGTGTAGGTCAGAG |
| Sequence-based reagent | mMFN2 R400Q knock-in mouse (reverse) | DornLab | Knock-in mice generated with point mutation in position 400 | CCCAGCTCCTCTGATTTGA |
| Sequence-based reagent | mMFN2 R400Q knock-in mouse (sequencing) | DornLab | Knock-in mice generated with point mutation in position 400 | CAGGTCTCCTTCCACCTTTAC |
| Sequence-based reagent | mMFN2 T105M knock-in mouse (forward) | DornLab | Knock-in mice generated with point mutation in position 105 | TGTTTACTTTGGAAGTAGGCAGTCT |
| Sequence-based reagent | mMFN2 T105M knock-in mouse (reverse) | DornLab | Knock-in mice generated with point mutation in position 105 | TTGTTCTTGGTGTCCCACTCTGA |
| Sequence-based reagent | mMFN2 T105M knock-in mouse (sequencing) | DornLab | Knock-in mice generated with point mutation in position 105 | TCGATGCTTAATGAGTGCTGCTGG |
| Sequence-based reagent | mMFN2 M376V knock-in mouse (forward) | DornLab | Knock-in mice generated with point mutation in position 376 | GTCCGGGCCAAGCAGATTGCAGAGGCCGTTCGTCTCATCATGGATTCCCTGCACATCGCAGCTCAGGAGCAGCGGTGAGA |
| Sequence-based reagent | mMFN2 M376V knock-in mouse (reverse) | DornLab | Knock-in mice generated with point mutation in position 376 | GGTAGTAAAGAGCCTTTCTAGCTGAT |
| Sequence-based reagent | mMFN2 M376V knock-in mouse (sequencing) | DornLab | Knock-in mice generated with point mutation in position 376 | TTTCGAGAGGCAGTTTGAGGTAA |
| Commercial assay or kit | GTPase-Glo Assay | Promega | Cat# V7681 | |
| Chemical compound, drug | Mitofusin agonist TAT-MP-1 | Thermo Fisher | Franco A et al., Nature 2016 | |
| Chemical compound, drug | Mitofusin antagonist TAT-MP-1 | Thermo Fisher | Franco A et al., Nature 2016 | |
| Chemical compound, drug | L-Glutamine | Gibco | Cat# 25030-149 | |
| Chemical compound, drug | Goat serum | Jackson ImmunoResearch | Cat# 005-000121 | |
| Chemical compound, drug | Doxorubicin | Sigma | 44583 | |
| Chemical compound, drug | Glutaraldehyde | Electron Microscopy Science | Cat# 16216 | |
| Chemical compound, drug | MitoTracker Green | Thermo Fisher | Cat# M7514 | |
| Chemical compound, drug | FCCP | Thermo Fisher | Cat# C2920 | |
| Chemical compound, drug | Calcein AM | Thermo Fisher | Cat# C3100MP | |
| Chemical compound, drug | Hematoxylin-eosin | Sigma | Cat#: GHS116 & HT110116 | |
| Chemical compound, drug | TUNEL | Promega | Cat#: G3250 | |
| Chemical compound, drug | Hoechst | Thermo Fisher | Cat# H3570 | |
| Chemical compound, drug | MitoTracker Orange | Thermo Fisher | Cat# M7510 | |
| Chemical compound, drug | Tetramethylrhodamine ethyl ester | Thermo Fisher | Cat# T-669 | |
| Chemical compound, drug | MitoSOX Red | Thermo Fisher | Cat# M36008 | |
| Software, algorithm | ImageJ | A. Schneider | https://imagej.net/Sholl_Analysis | |
| Software, algorithm | Partek Flow | https://www.partek.com/partek-flow/ | N/A | |
| Software, algorithm | FlowJo 10 software | https://www.flowjo.com/solutions/flowjo/downloads/ | N/A |
| MFN2 mutant | GTPase activity | Conformation change | Fusion | Polarization | Respiration | Mitophagy | Motility | KI phenotype |
|---|---|---|---|---|---|---|---|---|
| T105M | Inactive | Normal | Dominant suppressor | Dominant suppressor | Dominant suppressor | Normal | Dominant suppressor | CMT2A (het) Emb let (homo) |
| M376A/V | Normal | Impaired | Functional null | Normal | Functional null | Normal | Normal | Normal |
| R400Q | Normal | Impaired | Dominant suppressor | Normal | Functional null | Dominant suppressor | Normal | Cardiomyopathy (homo) |





