Hippo pathway-mediated YAP1/TAZ inhibition is essential for proper pancreatic endocrine specification and differentiation
Figures
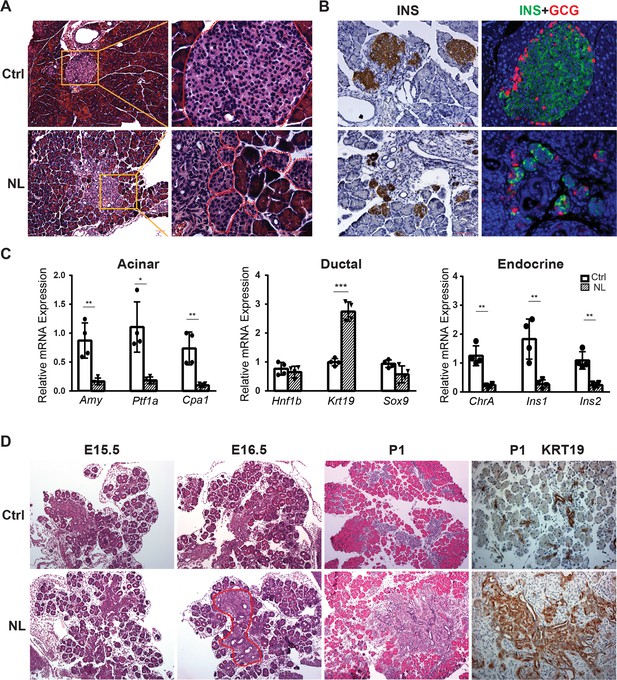
Deletion of Lats1/2 by Neurog3Cre perturbs pancreatic endocrine development.
(A) H&E staining of pancreatic tissue from NL mice showed phenotypically abnormal pancreatic islets as compared to those in control mice. (B) Immunostaining of Insulin (INS) and Glucagon (GCG) showed reduced size of pancreatic islets and decreased abundance of β-cells in NL mice. (C) Quantitative reverse transcription real-time PCR (RT-PCR) analysis of the expression levels of acinar, ductal, and endocrine genes in neonatal (P1) control and NL pancreas (Ctrl, n = 4 biological replicates; NL, n = 4 biological replicates). *p < 0.05; **p < 0.01; ***p < 0.001. (D) H&E staining and immunohistochemistry staining of KRT19 of pancreatic tissue from control and NL mice at indicated stage. Structural changes in histology and a higher level of KRT19 expression in NL mice were noted at the indicated stages. Scale bar: 100 μm.
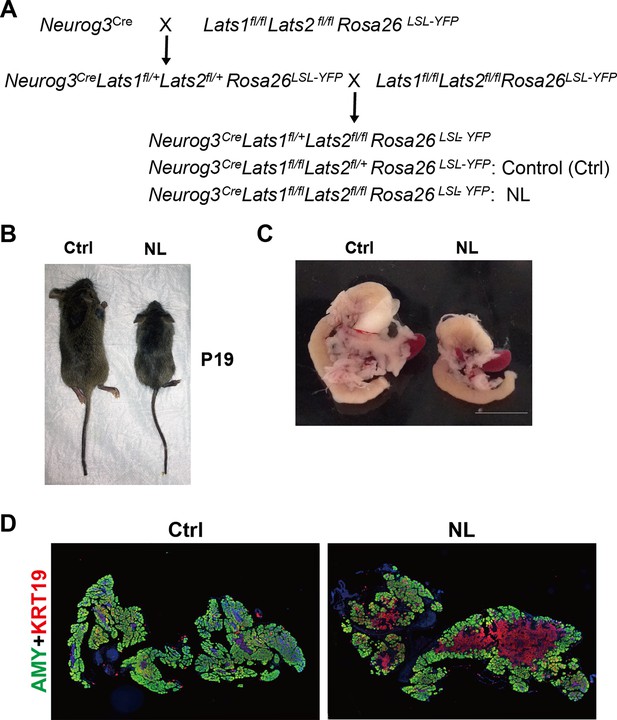
Deletion of Lats1/2 by Neurog3Cre perturbed pancreas differentiation.
(A) Schematic mating strategy of generating NL mice. (B) NL mice were associated with smaller body sizes as compared to the littermate controls at P19. (C) Macroscopic image of dissected P1 pancreas from control and NL mice. (D) Architectural changes of NL pancreas at P1 were determined by immunostaining with Amylase (AMY) and KRT19.
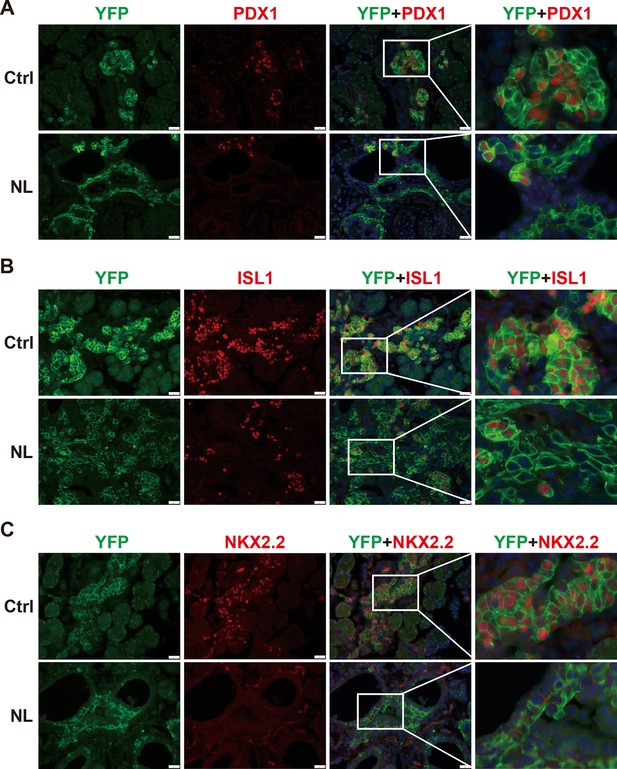
The differentiation of endocrine cells was blocked in NL pancreas at E16.5.
(A) Immunostaining of YFP and PDX1 on pancreatic tissue demonstrated that most of Lats1/2 null YFP+ cells in NL mice were negative for PDX1 staining. Immunostaining of YFP, Islet 1 (ISL1) (B), and NKX2.2 (C) showed that ISL1 and NKX2.2 were not expressed in the majority of YFP+ cells in NL pancreas. Scale bar: 50 μm.
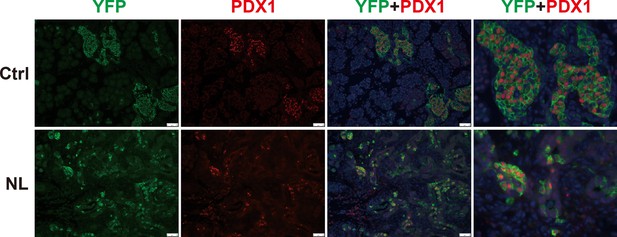
The differentiation of endocrine cells was blocked in NL pancreas at P1.
PDX1 was only expressed in a small fraction of YFP+ cells in the NL pancreas.
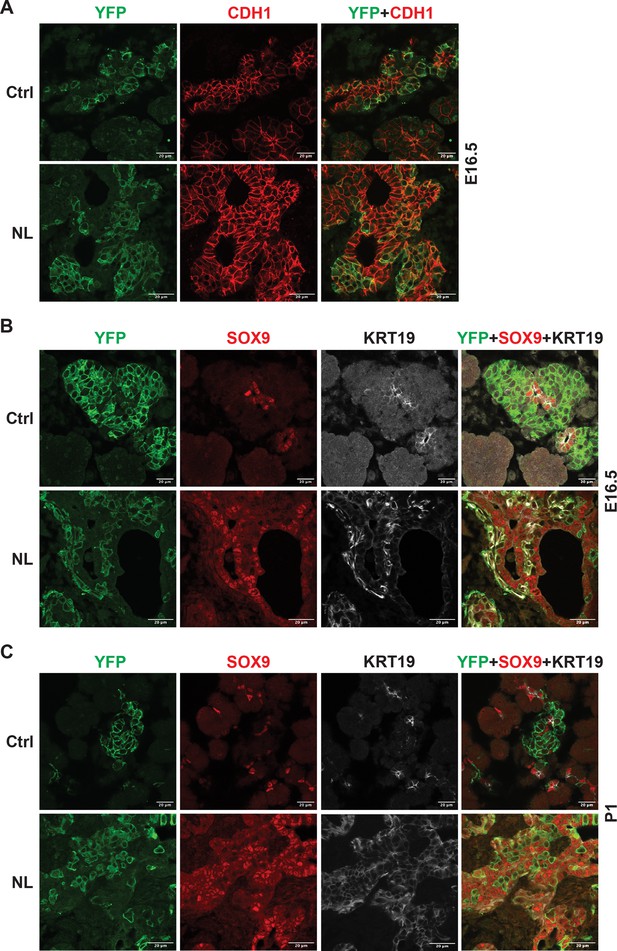
Loss of Lats1&2 altered endocrine progenitor cell characteristics.
(A) CDH1 protein expression in YFP+ cells was low in control pancreas and considerably higher in NL pancreas at E16.5. (B) In both control and NL pancreas, the YFP+ cells were negative for SOX9 staining at E16.5 and P1. (C) KRT19 staining was negative in YFP+ cells in control pancreas but appeared to be positive in YFP+ cells of NL pancreas at both E16.5 and P1. Scale bar: 20 μm.
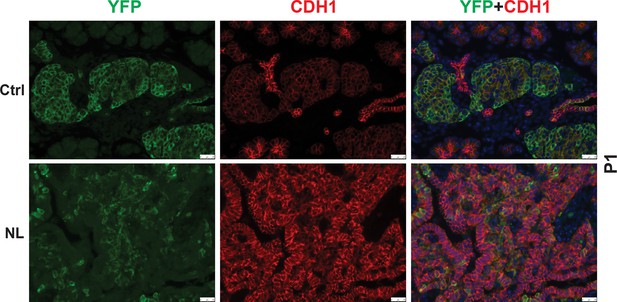
CDH1 expression in YFP+ cells was low in control pancreas and considerably higher in NL pancreas at P1.
Scale bar: 25 μm.
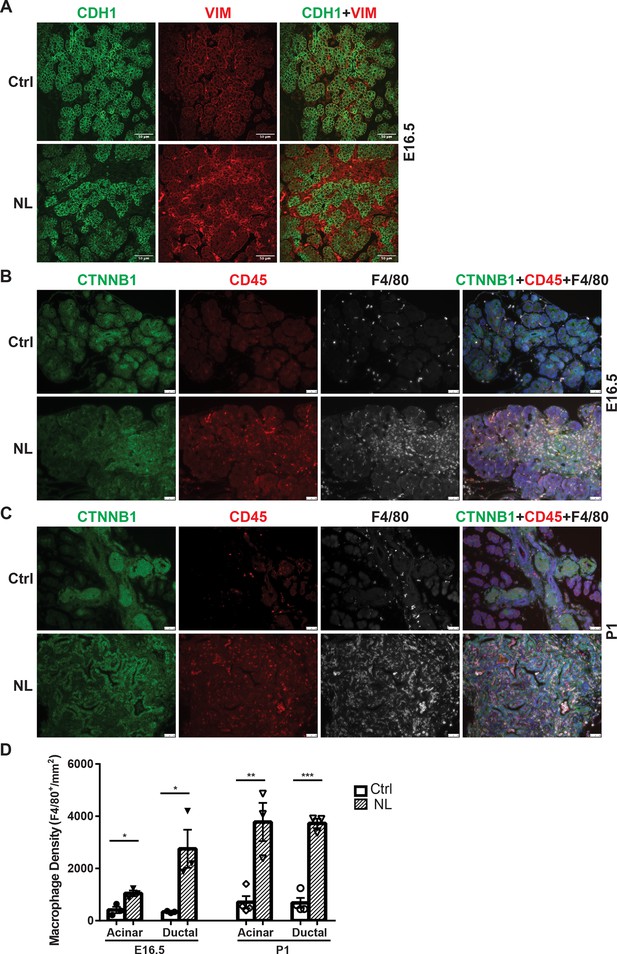
Loss of Lats1&2 in endocrine progenitor cells was associated with an increased number of mesenchymal cells and macrophages.
(A) Vimentin-positive staining was increased in NL pancreas at E16.5. Scale bar: 25 μm. Immunostaining showed that the number of CD45-positive immune cells and F4/80-positive macrophages was significantly increased in NL pancreas at E16.5 (B) and became even more apparent at P1 (C). (D) Quantification of F4/80+ macrophage showed a significant increase in NL pancreas compared to control pancreas at both E16.5 and P1 (E16.5: Ctrl, n = 3 biological replicates; NL, n = 3 biological replicates) (P1: Ctrl, n = 4 biological replicates; NL, n = 3 biological replicates). *p < 0.05; **p < 0.01; ***p < 0.001. Scale bar: 50 μm.
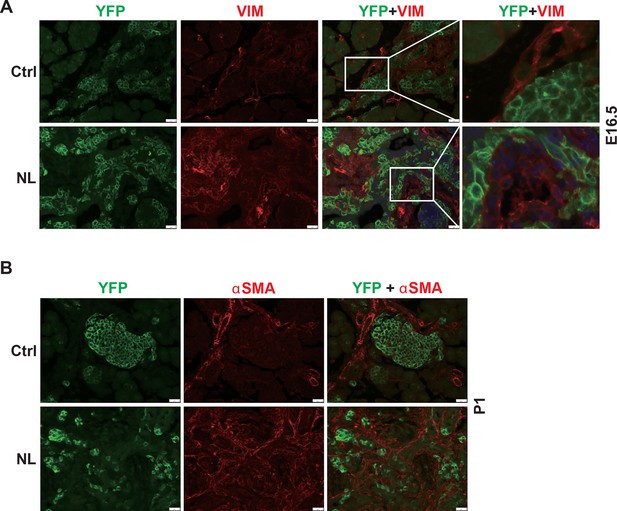
Loss of Lats1&2 in endocrine progenitor cells was associated with increased mesenchymal cells.
(A) Immunostaining of YFP and Vimentin revealed the expansion of mesenchymal cells next to YFP+ Lats1&2 null cells at E16.5. (B) The mesenchymal cells were positive for αSMA in P1 NL pancreas. Scale bar: 25 μm.
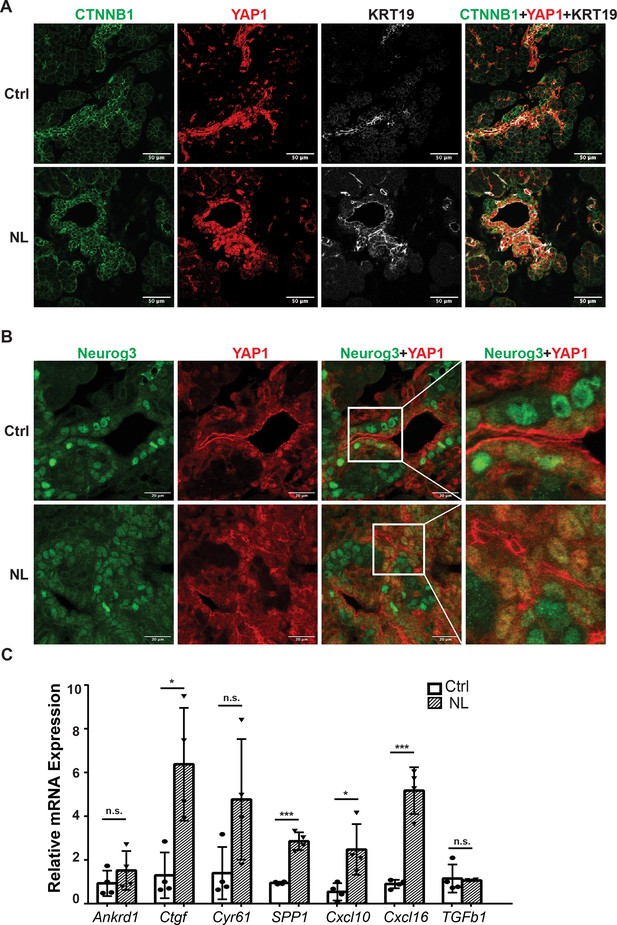
The expressions of YAP1/TAZ proteins and their targets were increased in Lats1&2 null cells.
(A) YAP1-positive cells showed increased CTNNB1 and KRT19 expression in both control and NL pancreases at E16.5. (B) YAP1 was sequestered to the cytoplasm in newly born Neurog3-positive cells located in the epithelial cord in control pancreas, while co-localization of YAP1 with Neurog3 was observed in nuclei of cord epithelial cells in NL pancreas at E16.5. (C) The mRNA levels of YAP1 targets Ctgf, SPP1, Cxcl10, and Cxcl16 were significantly increased in NL pancreas at P1 (Ctrl, n = 4 biological replicates; NL, n = 4 biological replicates). *p < 0.05; ***p < 0.001. Scale bar: 50 μm (A) and 20 μm (B).
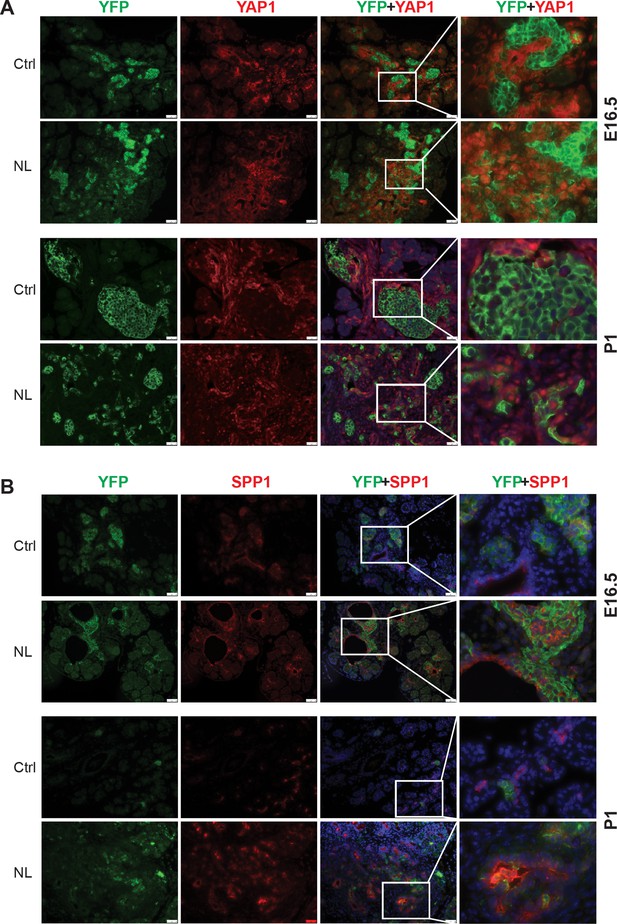
YAP1 and its targets were increased in Lats1&2 null cells.
(A) YAP1 was not expressed in YFP+ endocrine cells in control pancreas, but was present in nuclei of YFP+ cells in NL pancreas at P1. (B) SPP1 was expressed in YFP+ cells in NL pancreas, but not in control at E16.5 and P1. Scale bar: 50 μm.
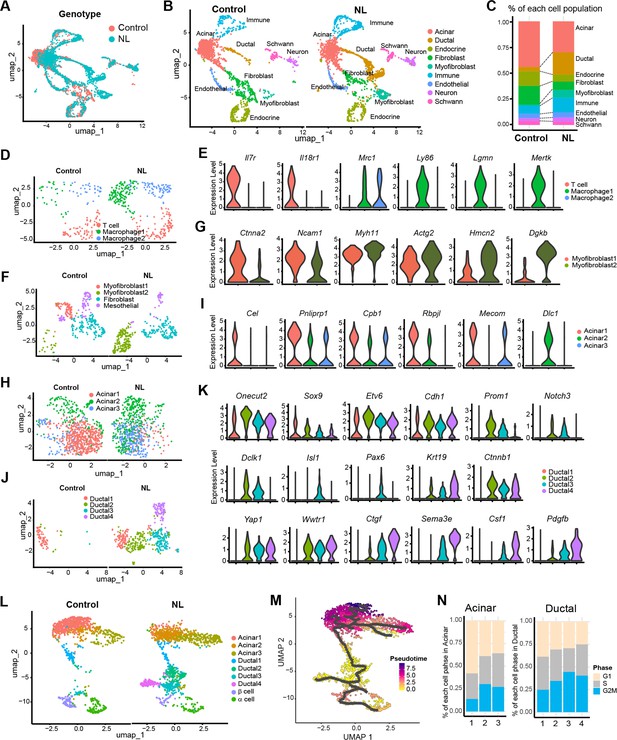
Single-nucleus RNA-seq analysis of control and NL mutant mice pancreas.
(A) Uniform manifold approximation and projection (UMAP) of all single nuclei present in control and NL mice pancreas that passed the QC steps. (B) UMAP with cell type annotation for each identified cluster in control and NL pancreas, respectively. (C) Percentage composition of each cell type in control and NL pancreas, respectively. (D) UMAP of sub-clustered immune cells in control and NL pancreas, respectively. (E) Violin plot of expression level of indicated genes in each sub-cluster shown in (D), with control and NL pancreas combined. (F) UMAP of sub-clustered fibroblasts and myofibroblasts in control and NL pancreas, respectively. (G) Violin plot of expression level of indicated genes in each sub-cluster shown in (F). (H) UMAP of sub-clustered acinar cells in control and NL pancreas, respectively. (I) Violin plot of expression level of indicated genes in each sub-cluster shown in (H). (J) UMAP of sub-clustered ductal cells in control and NL pancreas, respectively. (K) Violin plot of expression level of indicated genes in each sub-cluster shown in (J). (L) UMAP of sub-clustered pancreatic cells (acinar, ductal, and endocrine) in control and NL pancreas, respectively. (M) UMAP of single-cell RNA trajectory across the cell clusters shown in (L), with control and NL pancreas combined. Ductal 2 cell cluster was set as the root. (N) Percentage of cells in G1, S, and G2M phase in each acinar and ductal sub-clusters (control and NL pancreas combined).
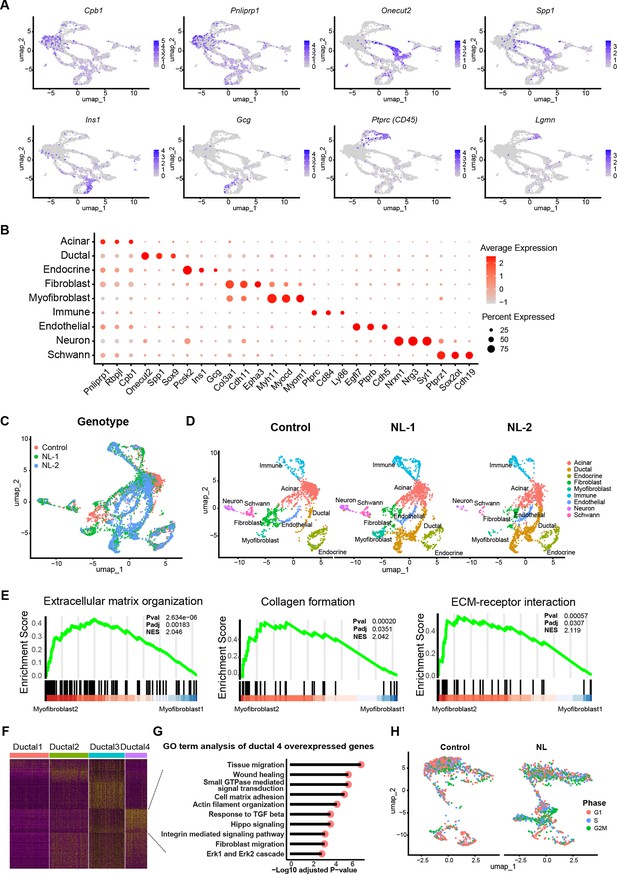
Single nuclei RNA-seq analysis of control and NL mutant mice pancreas.
(A) Uniform manifold approximation and projection (UMAP) of all single nuclei present in 1 control and 2 NL mice pancreas that passed the QC steps. (B) UMAP with cell type annotation for each identified cluster in 1 control and 2 NL pancreas, respectively. (C) Expression pattern of indicated genes in all single nuclei with control and NL pancreas combined. (D) Dot plot of indicated gene expression in each cell type present in the combined dataset. The color represents the average gene expression level in all cells within a cell type, the dot size represents the percentage of cells within a cell type that express indicated genes. (E) GSEA analysis of myofibroblast 1 and 2. (F) Heatmap of gene expression patterns of the differentially expressed genes (DEGs) across all four Ductal sub-clusters. The DEGs were identified by pairwise comparing each of the four sub-clusters. The resulting genes were filtered to only include DEGs with log2 fold change >1.5 or <−1.5, and adjusted p < 0.01. (G) Results of Gene Ontology analysis of genes that are highly expressed in Ductal 4. (H) UMAP of sub-clustered pancreatic cells (acinar, ductal, and endocrine) in control and NL pancreas, respectively. The cells were colored based on their status in different cell cycle phases.
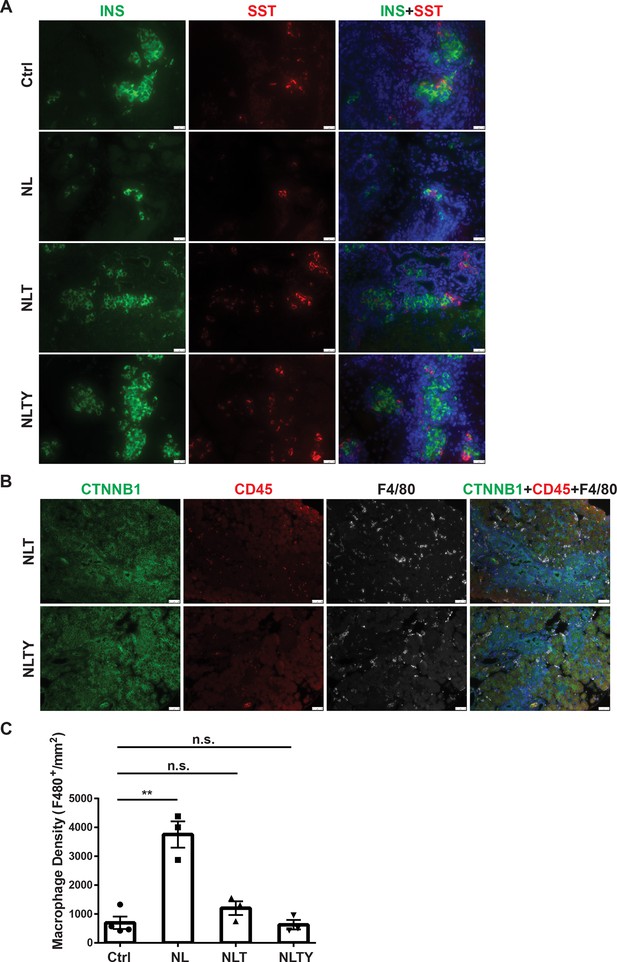
Removal of Yap1/Wwtr1 rescued the defect in endocrine specification and differentiation in NL pancreas.
(A) Co-staining of Insulin (INS) and Somatostatin (SST) showed that only small cell clusters were positive for INS or SST in NL pancreas, whereas the islets in NLT and NLTY pancreases were INS- or SST-positive, more closely resembling the control islets. Scale bar: 25 μm. (B) The numbers of immune cells were much less in both NLT and NLTY pancreases compared to NL pancreas. (C) Quantification analysis showed that while the macrophage density of NL pancreas was significantly higher compared to control pancreas, there was no significant difference in macrophage density between control and both NLT and NLTY pancreases (Ctrl, n = 4 biological replicates; NL, n = 3 biological replicates; NLT, n = 3 biological replicates; NLTY, n = 3 biological replicates). n.s. p > 0.05; **p < 0.01. Scale bar: 50 μm.
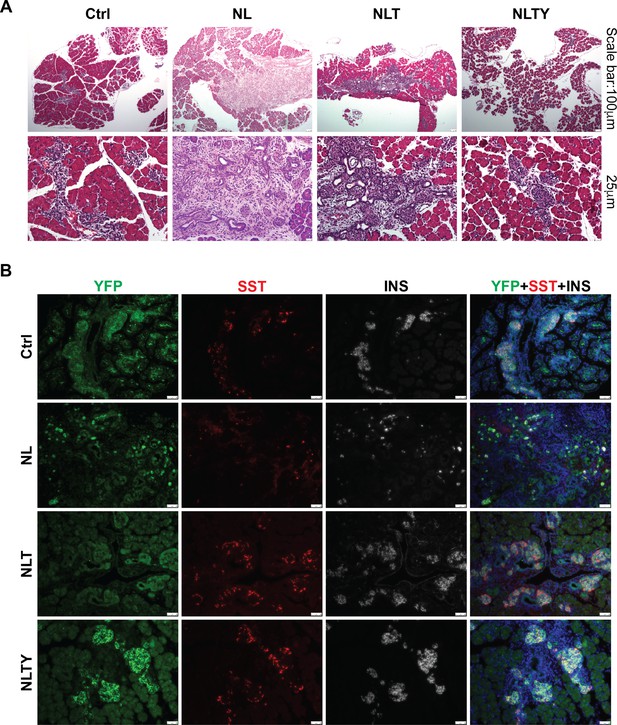
Removal of YAP1/TAZ rescued the defect in endocrine differentiation in NL pancreas.
(A) H&E staining showed morphologic defect in NLT pancreas, but appeared normal in NLTY pancreas. (B) Co-staining of YFP, Insulin (INS), and Somatostatin (SST) showed that only a small fraction of YFP+ cells were also positive for INS or SST in NL pancreas, whereas most YFP+ cells in NLT and NLTY pancreas were INS or SST positive, more closely resembling the control phenotype.
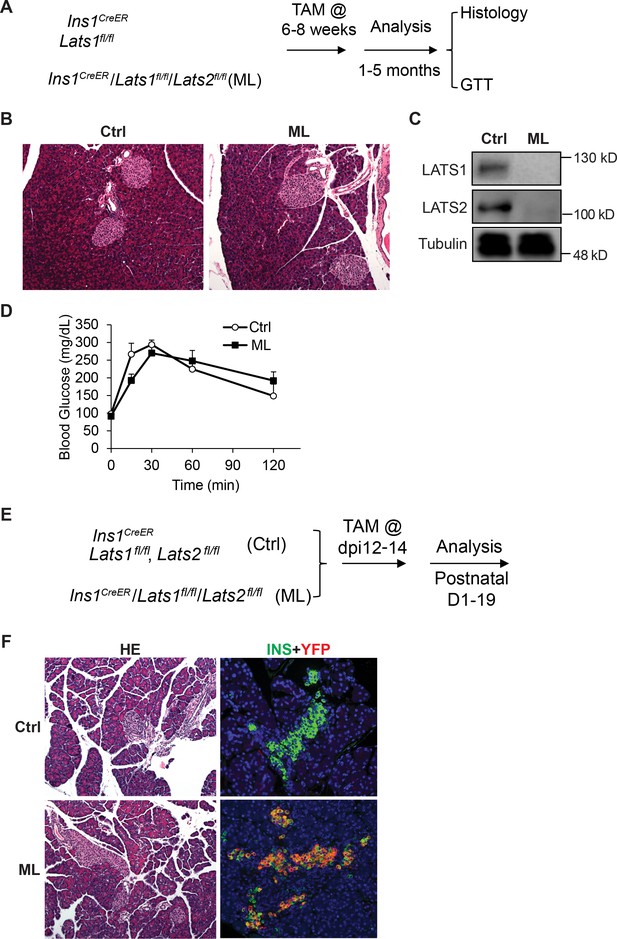
Lats1&2 are dispensable for pancreas β-cell proliferation and function.
(A) Schematic strategy of deleting Lats1&2 in adult β-cells using Ins1CreER to generate ML mice. (B) H&E staining showed normal pancreatic architecture and similar pancreatic islet size between control and ML pancreases. (C) LATS1&2 protein levels were reduced in pancreatic islets of ML mice. (D) The ML mice showed a normal glucose tolerance test (GTT) compared with control mice (Ctrl, n = 6 biological replicates; ML, n = 5 biological replicates). (E) Schematic strategy of deleting Lats1&2 in embryonic β-cells. (F) H&E staining showed normal pancreas structure between control and ML mice. Most YFP+ cells expressed INS in P1 ML pancreas.
-
Figure 8—source data 1
The original raw unedited blots for LATS1, LATS2, and TUBULIN demonstrated in Figure 8, as well as a figure of the uncropped blots with the relevant bands clearly labeled.
- https://cdn.elifesciences.org/articles/84532/elife-84532-fig8-data1-v2.zip
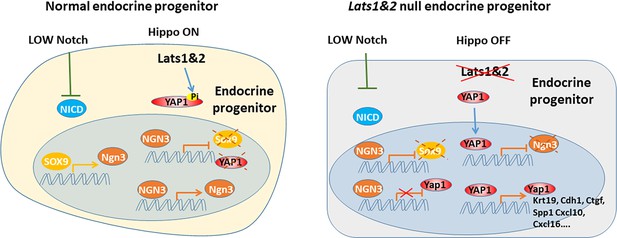
During endocrine progenitor specification, the Hippo pathway is required to sequester YAP1 protein in the cytosol and allow Neurog3 protein to positively regulate its own expression and suppress Sox9 and Yap1 expression.
Loss of Lats1&2 leads to YAP1 activation which suppresses Neurog3 expression and induces the expression of YAP1 targets.
Tables
Primers for genotyping and qRT-PCR.
| Gene name | Forward | Reverse |
|---|---|---|
| Lats1 | GCGATGTCTAGCCCATTCTC | GGTTGTCCCACCAACATTTC |
| Lats2 | AGCCTGACAACATACTCATCG | AATCCAGTGCAGAGGCCAAA |
| Yap1 | TACTGATGCAGGTACTGCGG | TCAGGGATCTCAAAGGAGGAC |
| Wwtr1 | GAAGGTGATGAATCAGCCTCTG | GTTCTGAGTCGGGTGGTTCTG |
| Ankrd1 | TAATCGCTCACAATCTGTTGACA | GCCTCTCACCTTCCGACCT |
| Ctgf | GGCCTCTTCTGCGATTTCG | GCAGCTTGACCCTTCTCGG |
| Cyr61 | TCCTCAGTGAGTTGCCCTC | CCCACCTAAGAGCCTCAGG |
| Spp1 | AGCAAGAAACTCTTCCAAGCAA | GTGAGATTCGTCAGATTCATCCG |
| Cxcl10 | CCAAGTGCTGCCGTCATTTTC | GGCTCGCAGGGATGATTTCAA |
| Cxcl16 | CCTTGTCTCTTGCGTTCTTCC | TCCAAAGTACCCTGCGGTATC |
| Tgfb1 | CCACCTGCAAGACCATCGAC | CTGGCGAGCCTTAGTTTGGAC |
| Amy | TTGCCAAGGAATGTGAGCGAT | CCAAGGTCTTGATGGGTTATGAA |
| Ptf1a | TCCCATCCCCTTACTTTGATGA | GTAGCAGTATTCGTGTAGCTGG |
| Cpa1 | CAGTCTTCGGCAATGAGAACT | GGGAAGGGCACTCGAACATC |
| Hnf1b | AGGGAGGTGGTCGATGTCA | TCTGGACTGTCTGGTTGAACT |
| Sox9 | AGTACCCGCATCTGCACAAC | ACGAAGGGTCTCTTCTCGCT |
| Krt19 | GGGGGTTCAGTACGCATTGG | GAGGACGAGGTCACGAAGC |
| Ins1 | CACTTCCTACCCCTGCTGG | ACCACAAAGATGCTGTTTGACA |
| Ins2 | GCTTCTTCTACACACCCATGTC | AGCACTGATCTACAATGCCAC |
| ChrA | ATCCTCTCTATCCTGCGACAC | GGGCTCTGGTTCTCAAACACT |
| GAPDH | AGGTCGGTGTGAACGGATTTG | GGGGTCGTTGATGGCAACA |
Primary antibodies list.
| Primary antibody | Company | Catalog # | Dilution | Application |
|---|---|---|---|---|
| alpha-Amylase (AMY) | Sigma-Aldrich | A8273 | 1:500 | IF |
| alpha-SMA (ACTA2) | Santa Cruz Biotechnology | 53142 | 1:250 | IF |
| beta-Catenin (CTNNB1) | BD Biosciences | 610153 | 1:100 | IF |
| CD45 | Biolegend | 103102 | 1:50 | IF |
| KRT19 | DSHB | Troma-III | 1:50 | IF, IHC |
| E-cadherin (CDH1) | Cell Signaling Technology | 3195S | 1:500 | IF |
| F4/80 | Cell Signaling Technology | 70076S | 1:50 | IF |
| GFP (YFP) | Aves | GFP-1020 | 1:500 | IF |
| Insulin (INS) | Abcam | ab7842 | 1:250 | IF |
| ISL1/2 | DSHB | 39.4D5 | 1:50 | IF |
| Ki67 | Biolegend | 652401 | 1:25 | IF |
| Neurogenin 3 (Neurog3) | DSHB | F25A1B3 | 1:50 | IF |
| PDX1 | DSHB | F109-D12 | 1:50 | IF |
| Somatostatin (SST) | Santa Cruz Biotechnology | sc-7819 | 1:100 | IF |
| SOX9 | Cell Signaling Technology | 82630S | 1:50 | IF |
| SPP1 | RD Systems | AF808 | 1:50 | IF |
| TAZ | Sigma-Aldrich | HPA007415-100UL | 1:50 | IF |
| YAP | Cell Signaling Technology | 14074S | 1:100 | IF |
| Tubulin | Proteintech | 11224-1-AP | 1:1000 | WB |
| LATS1 (C66B5) | Cell Signaling Technology | 3477 | 1:1000 | WB |
| LATS2 | Bethyl Laboratories, Inc | A300-479A | 1:1000 | WB |
Secondary antibodies list.
| Secondary antibody | Company | Catalog # | Dilution | Application |
|---|---|---|---|---|
| Alexa Fluor 488-conjugated AffiniPure Donkey Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | 715-545-150 | 1:250 | IF |
| Alexa Fluor 647-conjugated AffiniPure Donkey Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | 715-605-150 | 1:250 | IF |
| Cy3-conjugated AffiniPure Donkey Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | 715-165-150 | 1:250 | IF |
| Alexa Fluor 647-conjugated AffiniPure Donkey Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | 711-605-152 | 1:250 | IF |
| Cy3-conjugated AffiniPure Donkey Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | 711-165-152 | 1:250 | IF |
| Alexa Fluor 647-conjugated AffiniPure Donkey Anti-Rat IgG (H+L) | Jackson ImmunoResearch | 712-605-150 | 1:250 | IF |
| Cy3-conjugated AffiniPure Donkey Anti-Rat IgG (H+L) | Jackson ImmunoResearch | 712-165-150 | 1:250 | IF |
| Alexa Fluor 647-conjugated AffiniPure Donkey Anti-Goat IgG (H+L) | Jackson ImmunoResearch | 705-605-147 | 1:250 | IF |
| Cy3-conjugated AffiniPure Donkey Anti-Goat IgG (H+L) | Jackson ImmunoResearch | 705-165-003 | 1:250 | IF |
| Alexa Fluor 647-conjugated AffiniPure Donkey Anti-Guinea Pig IgG (H+L) | Jackson ImmunoResearch | 706-605-148 | 1:250 | IF |
| Alexa Fluor 488-conjugated AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) | Jackson ImmunoResearch | 703-545-155 | 1:250 | IF |
| Peroxidase-conjugated AffiniPure Goat Anti-Rat IgG (H+L) | Jackson ImmunoResearch | 112-035-003 | 1:250 | IHC |
| Peroxidase-conjugated AffiniPure Goat Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | 111-035-003 | 1:5000 | WB |





