Inhibition of type I PRMTs reforms muscle stem cell identity enhancing their therapeutic capacity
Figures
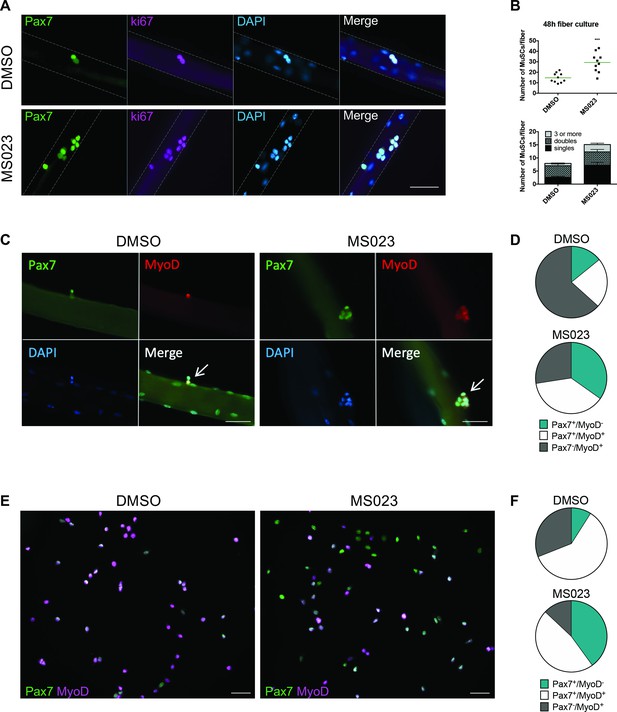
Enhanced self-renewal of MS023-treated MuSCs.
(A) Muscle fibers cultured for 48 hr with MS023 or DMSO. Pax7 identifies MuSCs, and ki67 identifies proliferating cells. Scale bar represents 50 μM. (B) Quantification of total MuSCs per fiber. Thirty fibers were counted per condition, horizontal bar represents average number of MuSCs per fiber (n=3 mice per condition). (C) Muscle fibers cultured for 48 hr with MS023 or DMSO. 30 fibers counted per condition (n=3 mice per condition). Scale bar represents 50 μM. (D) Quantification of Pax7/MyoD expressing MuSCs from (C). (E) Pax7/MyoD immunofluorescence staining of cultured myoblasts treated with MS023 or DMSO for 48 hr. Scale bar represents 50 μM. (F) Quantification of Pax7/MyoD expressing cells from (E).
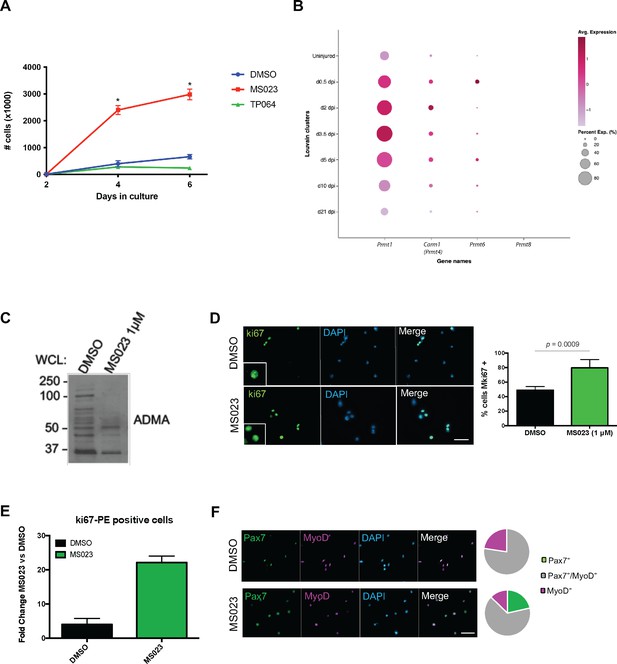
Type I PRMT inhibitor MS023 increases proliferation of MuSCs most by inhibition of PRMT1.
(A) Primary MuSCs were isolated from WT mice and seeded on collagen-coated 6 well plates. Upon seeding, cells were treated with 1 μM MS023, 1 μM TP064, or DMSO for 6 days. Cells were trypsinized and counted at days 2, 4, and 6 using a haemocytometer. (B) We investigated their expression from publicly available single cell RNAseq dataset (Oprescu et al., 2020), which performed analysis on skeletal muscle at different time points post-cardiotoxin injury (uninjured, and 0.5, 2, 3.5, 5, 10, 21 days post-injury). (C) Western blot of total asymmetric dimethyl arginine (ADMA) from whole cell lysate of primary myoblasts treated with MS023 (1 μM) or DMSO. (D) Ki67 immunofluorescence of MS023-treated primary myoblasts. Error bars represent mean ± SEM from three individual experiments, minimum of 200 cells per condition counted. Scale bar represents 50 μM. (E) Ki67-PE immunostained MS023-treated primary myoblasts analyzed with FACS, foldchange of mean intensity calculated compared to DMSO-treated cells. Error bars represent mean ± SEM from three individual replicates. (F) (left) Immunofluorescence of 14th passage MuSCs with DMSO or MS023 treatment, Pax7 (green), MyoD (purple), counterstained with DAPI. Scale bar represents 50 μM. (right) Pie chart representing quantification of Pax7 and MyoD expression from cells in left panel.
-
Figure 1—figure supplement 1—source data 1
Uncropped immunoblot.
- https://cdn.elifesciences.org/articles/84570/elife-84570-fig1-figsupp1-data1-v1.zip
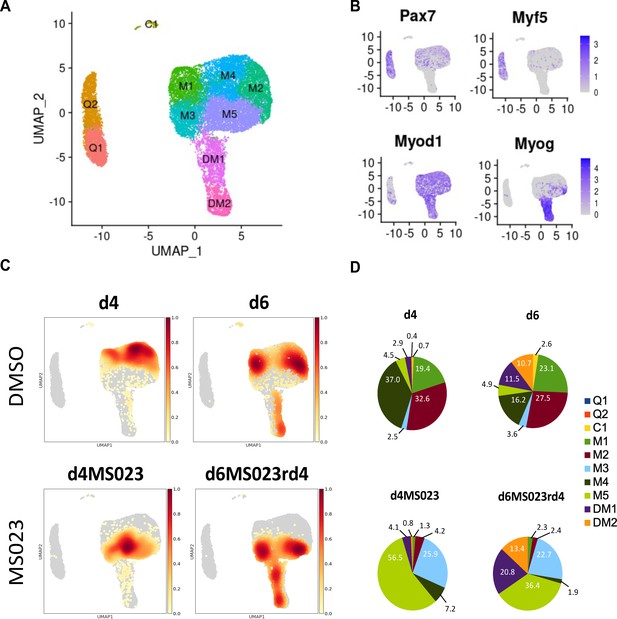
Single-cell graph-based clustering analysis.
(A) UMAP embedding representation for all cells. (B) Gene expression density on the UMAP embedding plot of myogenic markers Pax7, Myf5, MyoD, and Myogenin. (C) Distribution of cells from each sample within the UMAP embedding representation. (D) Proportion of cells from each sample belonging to each of the 10 identified clusters.
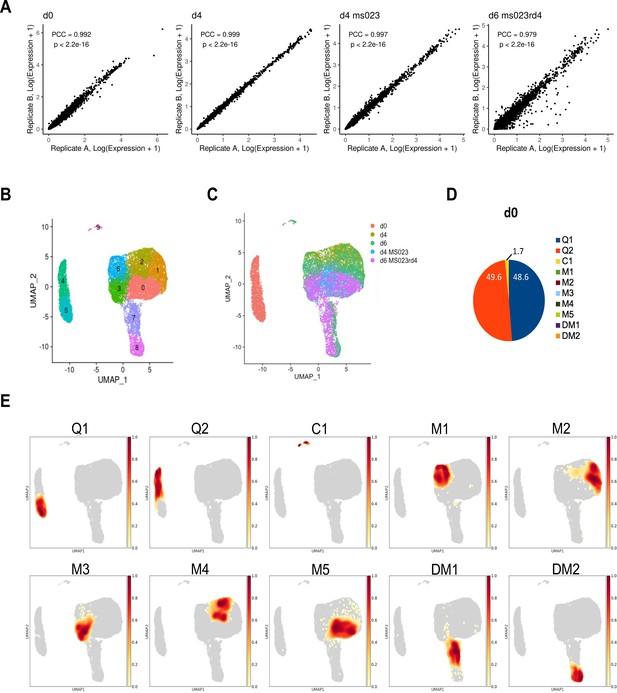
scRNAseq analysis of DMSO- and MS023-treated MuSCs.
(A) Reproducibility of average gene expression across biological replicates represented as Pearson correlation coefficients. (B) UMAP plot labelled with clusters 0–9. (C) Distribution of individual samples mapped onto the UMAP embedding representation for all cells. (D) Distribution of cells from d0 across the clusters represented in a pie chart. Cluster colour legend indicated on the right. (E) Distribution of individual clusters mapped onto the UMAP embedding representation for all cells.
scRNAseq analysis of DMSO- and MS023-treated MuSCs.
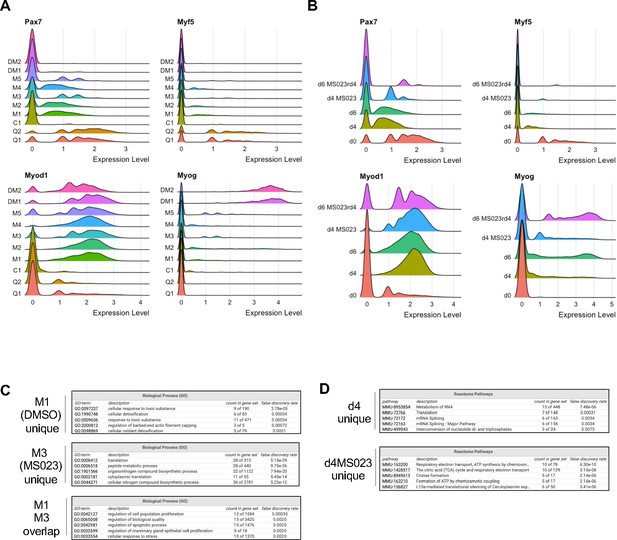
(A) Expression RidgePlots of key myogenic markers Pax7, Myf5, MyoD (Myod1) and Myog separated by cluster. (B) Expression RidgePlots of key myogenic markers Pax7, Myf5, MyoD (Myod1) and Myog separated by sample. (C) GO enrichment terms of top 100 marker genes unique to cluster M1 (DMSO specific, top panel), M3 (MS023 specific, middle panel), and genes that are common between clusters M1 (DMSO) and M3 (MS023) (lower panel). (D) GO enrichment terms of top 100 marker genes unique to d4 cells (top panel) and d4MS023 cells (lower panel).
scRNAseq analysis of DMSO- and MS023-treated MuSCs.
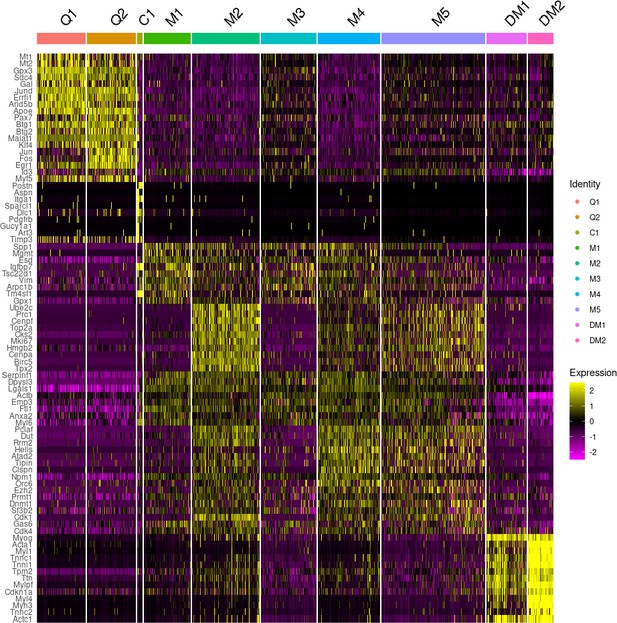
Heatmap of the top 100 enriched genes in each cluster.
Pseudotime trajectory of pooled DMSO- and MS023-treated MuSCs.
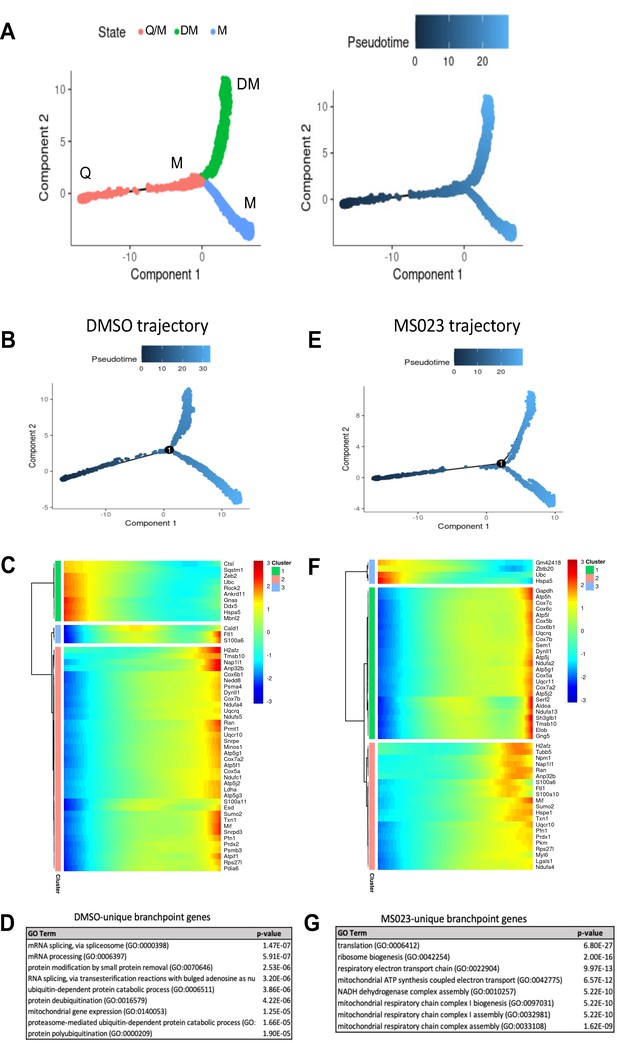
(A) Trajectory of all cells depicting three cell states. Pseudotime starts at the left endpoint of the plot. Q: Quiescence. M: Proliferating myoblast. DM: Differentiating myoblast. (B–D) Monocle trajectory of day-0 and DMSO-treated samples (B), heatmap of top differentially regulated genes across pseudotime (C) and GO enrichment analyses of DMSO-unique branchpoint genes (D). (E–G) Monocle trajectory of day-0 and MS023-treated samples (E), heatmap of top differentially regulated genes across pseudotime (F) and GO enrichment analyses of MS023-unique branchpoint genes (G).
Monocle trajectory analysis of DMSO- and MS023-treated MuSCs.
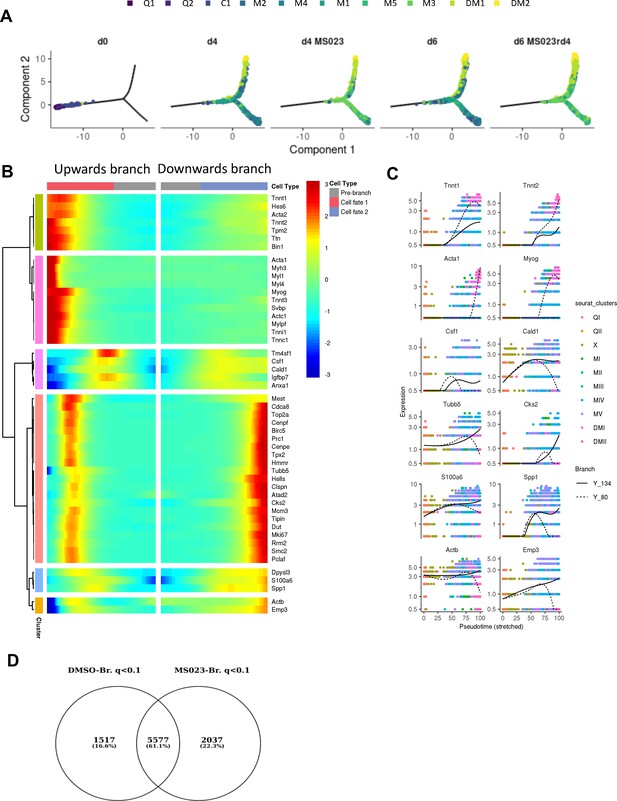
(A) Cell trajectories for each sample and cluster, with the pseudotime starting at the left endpoint of the plot. (B) Heatmap of significantly differentially expressed genes across the branchpoint of the pooled sample trajectory. (C) Distributions in the kinetic trends of each lineage for the expression for selected genes. (D) Venn diagram of overlapping differentially regulated genes across the branchpoint of the DMSO and MS023 trajectories with a q value <0.1.
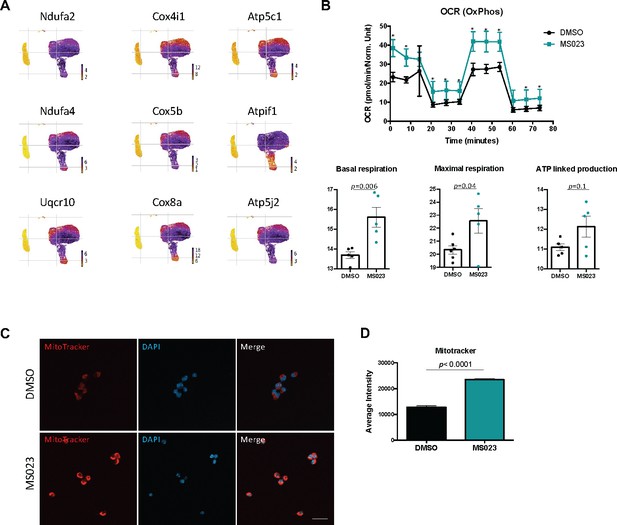
MS023-treated proliferating MuSCs display elevated oxidative phosphorylation.
(A) UMAP plots showing expression of components of the electron transport chain localized to MS023 clusters. (B) Seahorse XF96 analysis of oxygen consumption rate (OCR) in DMSO and MS023-treated freshly isolated primary MuSCs and quantification of basal respiration, maximal respiration, and ATP-linked production. Error bars represent mean ± SEM from seven individual wells per condition. (C) MitoTracker staining (red) of freshly isolated MuSCs treated with DMSO (upper panel) or MS023 (lower panel) and counterstained with DAPI to visualize nuclei. Minimum 100 cells quantified per condition from three independent experiments. Scale bar represents 20 μM. (D) Quantification of MitoTracker signal intensity of cells from (C), error bars represent mean ± SEM from three individual replicates, >100 nuclei quantified per condition (p<0.0001).
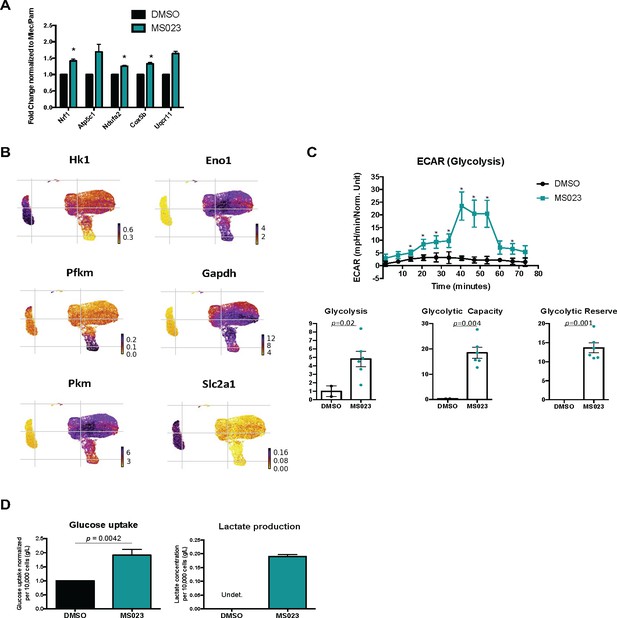
Glycolysis is increased MS023-treated MuSCs.
(A) Fold change of selected OxPhos genes in response to treatment with MS023. Bulk RNA was isolated from freshly purified primary myoblasts. Error bars represent mean ± SEM from 3 biological replicates. (B) UMAP plots showing expression of components of glycolysis localized to MS023 clusters. (C) Seahorse XF96 analysis of extracellular acidification rate (ECAR) in DMSO and MS023-treated freshly isolated primary MuSCs and quantification of glycolysis, glycolytic capacity, and glycolytic reserve. Error bars represent mean ± SEM from seven individual wells per condition. (D) Measurement of glucose uptake (left) and lactate production (right) as quantified from the media of d4 (DMSO) and d4MS023 (MS023) MuSCs. Error bars represent mean ± SEM from three independent experiments.
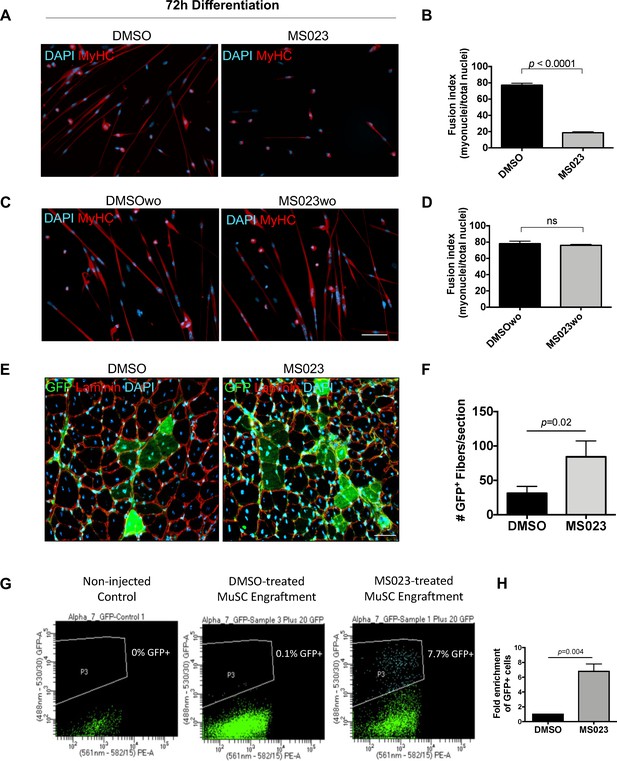
MS023 treatment is reversible and allows expanded MuSCs to differentiate ex vivo and in vivo.
(A) Immunofluorescence of MyHC counterstained with DAPI in primary myotubes that were differentiated in the presence of DMSO or MS023 following 4 days of treatment with DMSO or MS023. (B) Fusion index calculated as the ratio of nuclei within multinucleated myotubes to total nuclei from >200 nuclei per condition, error bars represent mean ± SEM from three individual replicates. (C) Immunofluorescence of MyHC counterstained with DAPI in primary myotubes that were differentiated in the absence of DMSO or MS023 following 4 days of treatment with DMSO or MS023 and 2 days of washout. Scale bar represents 50 μM (D) Fusion index calculated the same as 6B. (E) Immunofluorescence of GFP+ myofibers following transplantation of 15,000 DMSO or MS023-treated MuSCs. Scale bar represents 50 μM. (F) Quantification of GFP+ myofibers. Error bars represent mean ± SEM from three individual biological replicates. (G) Representative FACS plots of re-isolated MuSCs following engraftment and muscle regeneration. P3 represents the population of purified GFP+ MuSCs. (H) Fold enrichment of MS023-treated MuSC re-isolation compared to DMSO. Error bars represent mean ± SEM from three individual biological replicates.
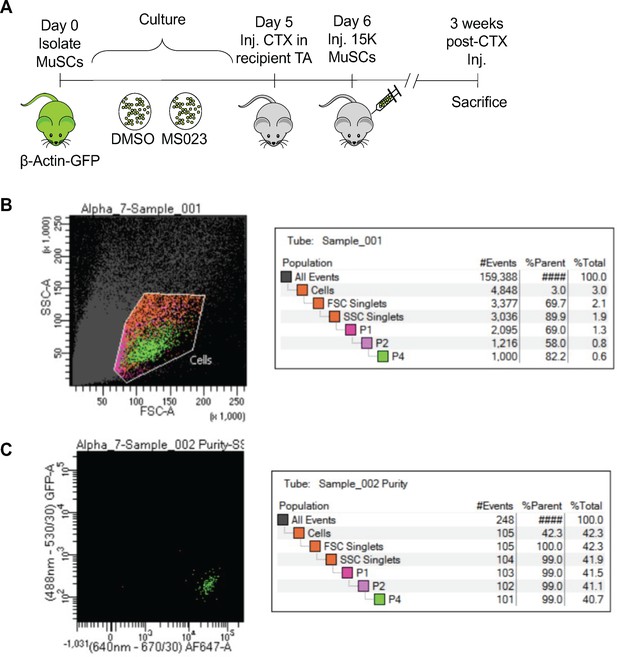
Type I PRMT inhibitor MS023 enhances MuSC engraftment.
(A) Experimental outline for MuSC engraftment. (B) FACS plot of primary purified MuSCs represented as P4 (0.6% of the total population). (C) The purity of sorted cells is 99%.
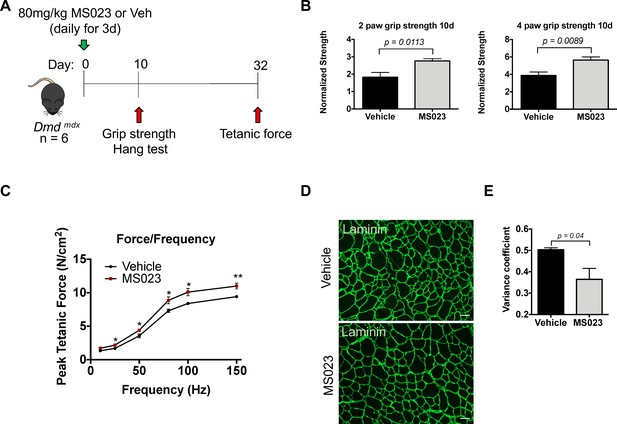
In vivo administration of MS023 to dystrophic mice improves muscle strength.
(A) Experimental schema. (B) Normalized grip strength measurements taken from two forelimbs and all four limbs of mice treated with vehicle or MS023 10 days after the last injection. Error bars represent mean ± SEM from six biological replicates. (C) Force/frequency curve generated for mice treated with vehicle or MS023. (D) Representative cross-sectional area of TA muscles isolated from mice treated with vehicle or MS023 and immunostained with anti-laminin antibodies to visualize myofibers. Scale bar represents 50 μM. (E) Quantification of the variance coefficient of TA muscle minimum fiber Feret measurement from mice treated with vehicle or MS023. Error bars represent mean ± SEM from three biological replicates, >200 fibers measured per mouse.
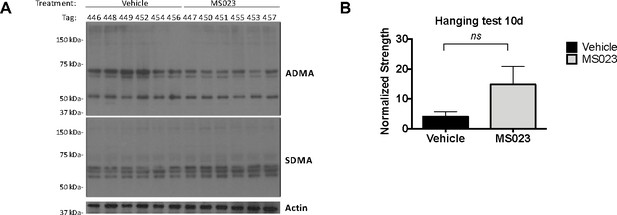
MS023 decreases ADMA containing proteins globally and increases strength in mdx mice.
(A) Whole tissue lysate from homogenized tail pieces of mice treated with Vehicle or MS023 probed with antibodies against total asymmetric dimethyl arginine (ADMA), symmetric dimethyl arginine (SDMA), and actin for loading control. The ADMA levels normalized to β−actin from left to right are: (0.94, 0.95, 1.03, 0.99, 0.91, 0.89), (0.86, 0.83, 0.83, 0.85, 0.78, 0.82). The SDMA levels normalized to β-actin from left to right are: (0.95, 0.89, 0.94, 0.90, 0.94, 0.91), (0.93, 0.95, 0.96, 0.93, 0.93, 0.94). (B) Hanging test measurements taken from mdx mice treated with vehicle or MS023 at 10d post-injection. Error bars represent mean ± SEM from six biological replicates.
-
Figure 6—figure supplement 1—source data 1
Uncropped immunoblot.
- https://cdn.elifesciences.org/articles/84570/elife-84570-fig6-figsupp1-data1-v1.zip
Additional files
-
Supplementary file 1
Expressed genes per cluster.
Quiescence 1 and 2 (Q1 and Q2), myoblast clusters 1–5 (M1-M5), and differentiated myoblasts 1 and 2 (DM1 and DM2) clusters.
- https://cdn.elifesciences.org/articles/84570/elife-84570-supp1-v1.xlsx
-
Supplementary file 2
Top 100 genes enriched per cluster of MuSCs purified from 8-week-old C57BL/6 mice immediately after (1) isolation (termed d0; sample day 0), and (2) culture in growth medium for 4 days with 0.033% DMSO as control (sample d4) or with 1 μM MS023 (sample d4MS023), and (3) grown in growth media for 6 days with 0.033% DMSO removed at day 4 (sample d6), or 6 days in culture with 1 μM MS023 removed at day 4 (sample d6MS023rd4).
- https://cdn.elifesciences.org/articles/84570/elife-84570-supp2-v1.xlsx
-
Supplementary file 3
Sample distribution across clusters.
The number of cells and the percentage distributed within clusters Q1, Q2, C1, M1, M2, M3, M4, M5, DM1 and DM2 is indicated.
- https://cdn.elifesciences.org/articles/84570/elife-84570-supp3-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/84570/elife-84570-mdarchecklist1-v1.docx





