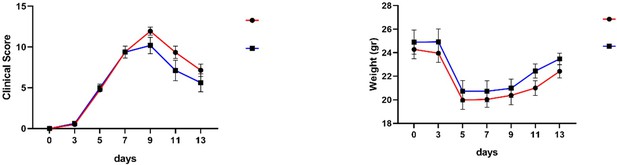
Mir221/222 drive synovial hyperplasia and arthritis by targeting cell cycle inhibitors and chromatin remodeling components
Figures
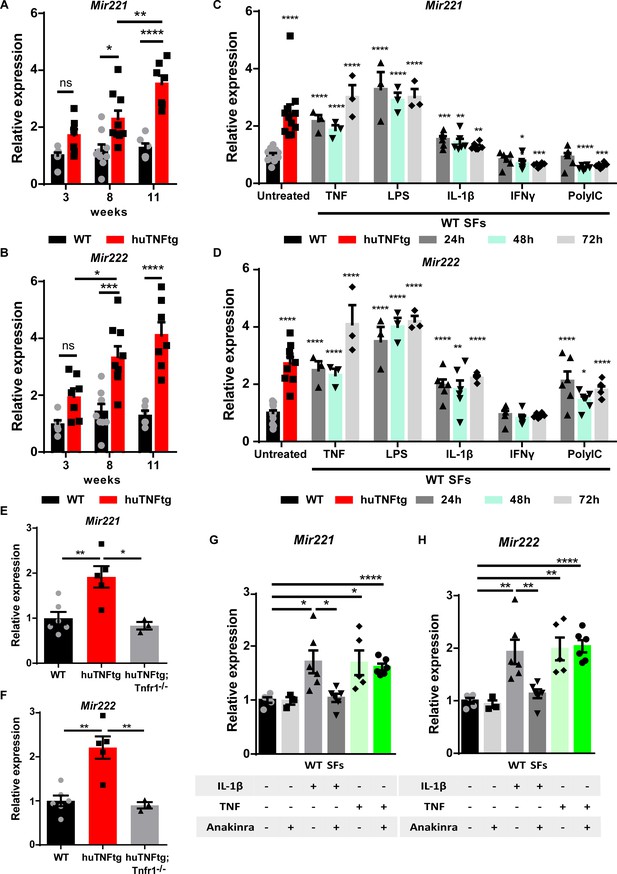
Regulation of Mir221/222 levels in synovial fibroblasts (SFs) under arthritogenic signals.
(A, B) Expression levels of Mir221 and Mir222 from cultured SFs, as determined by qRT-PCR at different time points of disease of huTNFtg mice, as well as control wild-type mice (WT). Expression levels were normalized to the levels seen in 3-week-old WT mice (n = 6–9). Two-way ANOVA statistical analysis was used with the suggested correction. (C, D) Quantification of Mir221 and Mir222 levels in cultured WT SFs after 24, 48, and 72 hr stimulation with TNF, LPS, IL1-β, IFN-γ, and PolyIC (n = 3–13). WT unstimulated SFs served as reference. Multiple t-tests statistical analysis was used. (E, F) Mir221 and 222 levels in cultured SFs from huTNFtg and huTNFtg;Tnfr1-/- mice (n = 3–6). WT SFs served as reference for normalization. Student’s t-test statistical analysis was used, unpaired. (G, H) Mir221 and 222 expression analysis in cultured WT SFs after TNF or IL-1β stimulation in the absence or presence of anakinra (n = 3–6). Expression was normalized to the levels detected in WT unstimulated SFs. Student’s t-test statistical analysis was used, unpaired. In all expression analysis experiments for Mir221 and 222, Rnu6 was used as a housekeeping gene. Data represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0,0001, ns = not significant.
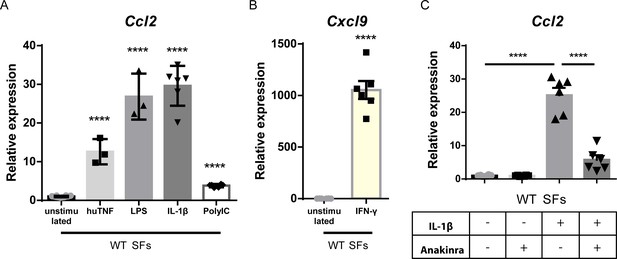
Synovial fibroblasts (SFs) respond to inflammatory signals.
(A) qRT-PCR analysis of Ccl2 expression in cultured WT SFs after 24 hr stimulation with TNF, LPS, IL1-β, and PolyIC (n = 3–6). WT unstimulated SFs served as reference. Student’s t-test statistical analysis was used, unpaired. (B) qRT-PCR analysis of Cxcl9 expression in cultured WT SFs after 24 hr stimulation with IFN-γ (n = 6). WT unstimulated SFs served as reference for normalization. Student’s t-test statistical analysis was used, unpaired. (C) qRT-PCR analysis of Ccl2 expression in cultured WT SFs after 24 hr stimulation with TNF and IL1-β in the presence or absence of anakinra (n = 3–6). Expression was normalized to the levels detected in WT unstimulated SFs. Student’s t-test statistical analysis was used, unpaired. In all expression analysis experiments, B2m was used as a housekeeping gene for normalization. Data represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0,0001, ns = not significant.
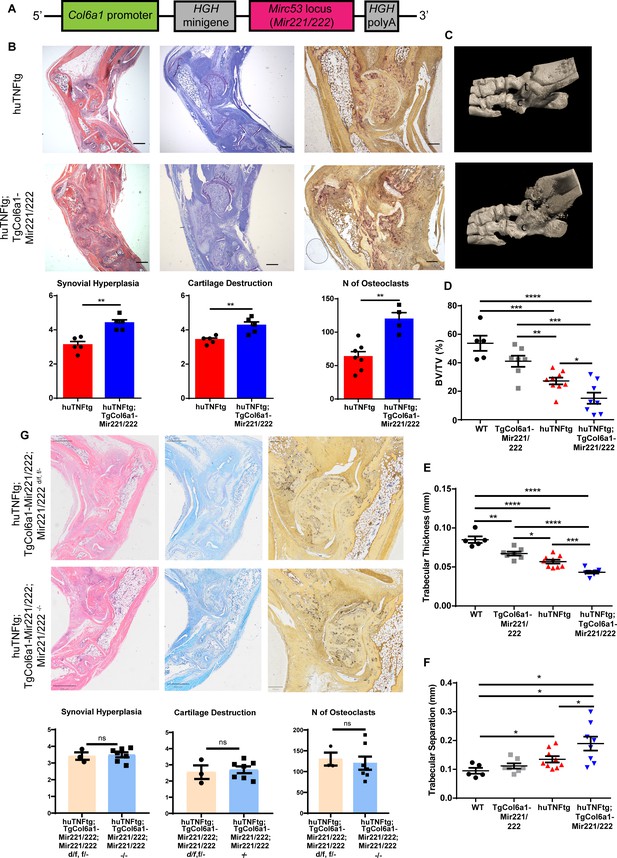
Mesenchymal Mir221/222 overexpression leads to worse arthritis manifestations in huTNFtg mice.
(A) Mir221/222 were cloned under the Col6α1 promoter to target expression in cells of mesenchymal origin. (B) Representative histological images of H&E, TB, and TRAP stained ankle joint sections and histological score of synovial hyperplasia, cartilage destruction, and osteoclast numbers of 8-week-old huTNFtg (n = 5–7) and huTNFtg;TgCol6a1-Mir221/222 mice (n = 4–5). t: talus; c: calcaneous. Scale bars: 600 μm and 300 μm. Student’s t-test statistical analysis was used, unpaired. (C) Representative microCT images of the ankle joint area of 8-week-old huTNFtg and huTNFtg;TgCol6a1-Mir221/222 mice. (D–F) Quantification of bone erosions measuring the following parameters: decreased bone volume, decreased trabecular thickness, and increased trabecular separation by microCT analysis in the ankle joints of WT (n = 5), TgCol6a1-Mir221/222 (n = 7), huTNFtg (n = 9), and huTNFtg;TgCol6a1-Mir221/222 mice (n = 8). BV: bone volume; TV: trabecular volume. Student’s t-test statistical analysis was used, unpaired. Data represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0,0001, ns = not significant. (G) Representative histological images of H&E, TB, and TRAP stained ankle joint sections and histological score of synovial hyperplasia, cartilage destruction, and osteoclast numbers of 7–8-week-old huTNFtg;TgCol6a1-Mir221/222;Mir221/222 d/f, f/- (n = 3) and huTNFtg;TgCol6a1-Mir221/222;Mir221/222-/- mice (n = 7). Scale bars: 800 μm and 250 μm. Student’s t-test statistical analysis was used, unpaired.
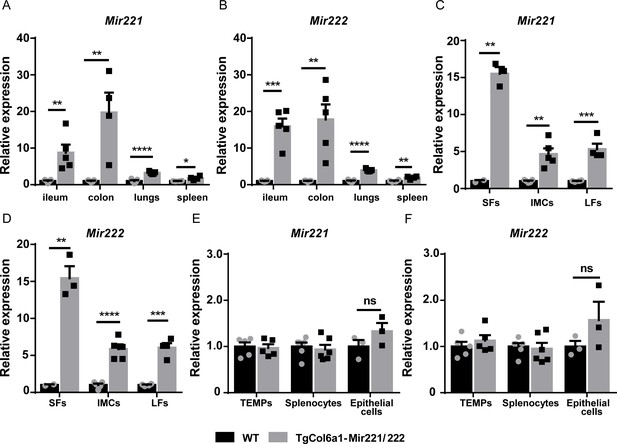
TgCol6a1-Mir221/222 mice target tissues and cells of mesenchymal origin.
Expression levels of Mir221 and Mir222 as determined by qRT-PCR from: (A, B) Ileum, colon, lung, and spleen from TgCol6a1-Mir221/222 mice. Student’s t-test statistical analysis was used, unpaired. (C, D) Synovial fibroblasts (SFs), intestinal mesenchymal cells (IMCs), and lung fibroblasts (LFs) from TgCol6a1-Mir221/222 mice. Student’s t-test statistical analysis was used, unpaired. (E, F) Peritoneal macrophages (TEMPs), splenocytes, and epithelial cells from TgCol6a1-Mir221/222 mice. Student’s t-test statistical analysis was used, unpaired. Expression levels were normalized to the levels seen in WT mice (n = 3–6). Rnu6 was used for normalization. Data represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0,0001, ns = not significant.
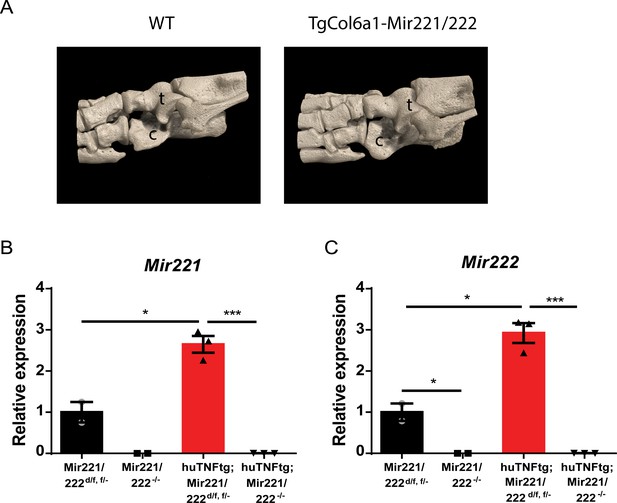
Mir221/222 overexpression may affect bone physiology. Synovial fibroblasts (SFs) from Mir221/222-/- mice lack Mir221/222 expression.
(A) Mir221/222 overexpression may affect bone physiology. Representative microCT images from WT and TgCol6a1-Mir221/222 ankle joint area. t: talus; c: calcaneous. (B, C) Synovial fibroblasts (SFs) from Mir221/222 -/- mice lack Mir221/222 expression. qRT-PCR for Mir221 and 222 levels in cultured SFs from 9 week-old Mir221/222 d/f, Mir221/222 -/-, huTNFtg;Mir221/222 d/f, and huTNFtg;Mir221/222 -/- (n = 2–3) mice. Expression of Mir221/222 d/f SFs was used for normalization. Student’s t-test statistical analysis was used, unpaired. In all experiments, Rnu6 was used as a housekeeping gene for normalization. Data represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0,0001, ns = not significant.
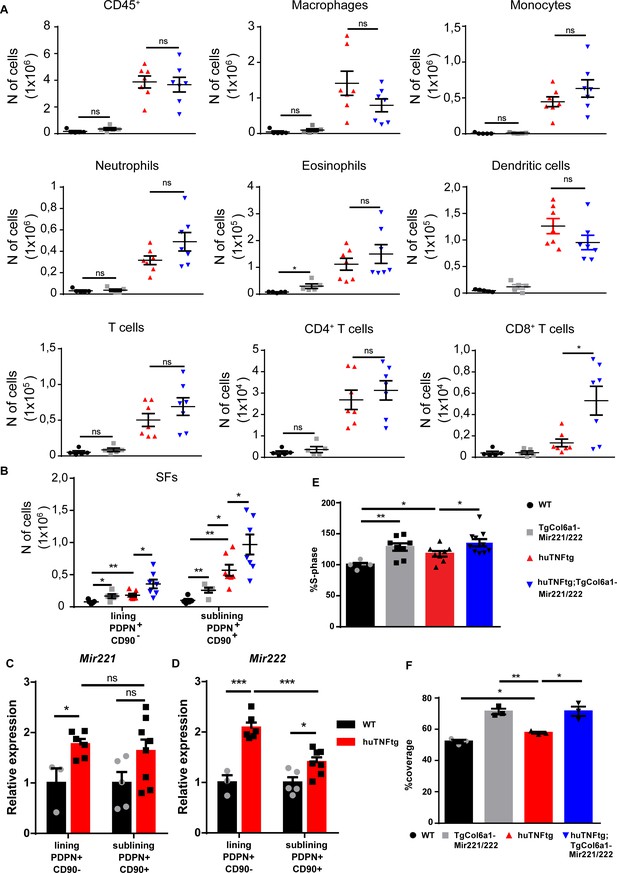
Mir221/222 overexpression leads to fibroblast expansion.
(A) Infiltration of CD45+ cells, macrophages, monocytes, neutrophils, eosinophils, dendritic cells, CD4+ T cells, and CD8+ T cells in the ankle joints of 8-week-old WT (n = 5), TgColVI-Mir221/222 (n = 5), huTNFtg (n = 7), and huTNFtg;TgCol6a1-Mir221/222 mice (n = 7) quantified by FACS analysis (from two independent experiments). Student’s t-test statistical analysis was used, unpaired. (B) Lining PDPN+CD90- and sublining PDPN+CD90+ synovial fibroblast (SF) number quantification in the ankle joints of 8-week-old WT (n = 5), TgCol6a1-Mir221/222 (n = 5), huTNFtg (n = 7), and huTNFtg;TgCol6a1-Mir221/222 mice (n = 7) by FACS analysis (from two independent experiments). Student’s t-test statistical analysis was used, unpaired. (C, D) Mir221 and 222 levels in freshly sorted lining PDPN+CD90- and sublining PDPN+CD90+ SFs in the ankle joints of 8-week-old WT and huTNFtg mice (n = 3–8). Student’s t-test statistical analysis was used, unpaired. Rnu6 was used as a housekeeping gene for normalization (E) % fraction of cultured SFs that are in the S-phase of the cell cycle from 8-week-old WT (n = 5), TgCol6a1-Mir221/222 (n = 8), huTNFtg (n = 8), and huTNFtg;TgCol6a1-Mir221/222 mice (n = 10) quantified by FACS analysis (from three independent experiments). Student’s t-test statistical analysis was used, unpaired. (F) % area that was covered by cultured fibroblasts 24 hr after a wound was performed (n = 3). A representative experiment out of three is presented. Student’s t-test statistical analysis was used, unpaired. All comparisons were performed using WT SFs as a reference sample. Data represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0,0001, ns = not significant.
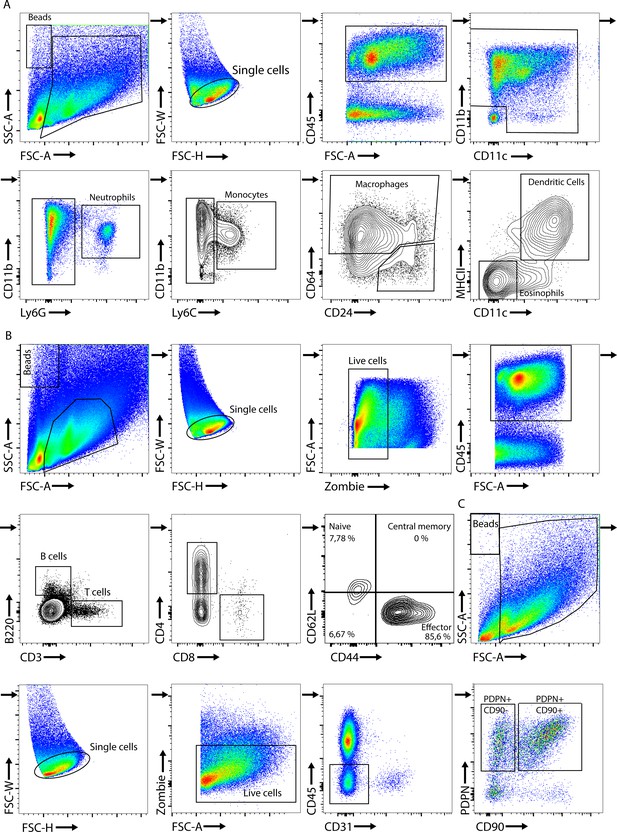
FACS gating strategies.
(A) FACS gating strategy for myeloid cell infiltration in the ankle joints. The cell suspension was gated for live CD45+. In the CD45+ population, we characterized the abundance of macrophages, monocytes, neutrophils, dendritic cells, and eosinophils. (B) FACS gating strategy for lymphocyte cell infiltration in the ankle joints. The cell suspension was gated for live CD45+, and we plotted for CD4+ and CD8+ T cells. Then, the CD8+ T cells were further characterized for CD62L and CD44 expression. (C) FACS gating strategy for synovial fibroblast populations in the ankle joints. The cell suspension was gated for live CD45-CD31-, and we gated for PDPN and CD90 expression on fibroblasts. Single-cell suspensions from ankle joints were gated for live cells using zombie green or NIR. Counting beads were used for quantification of different cell subpopulations.
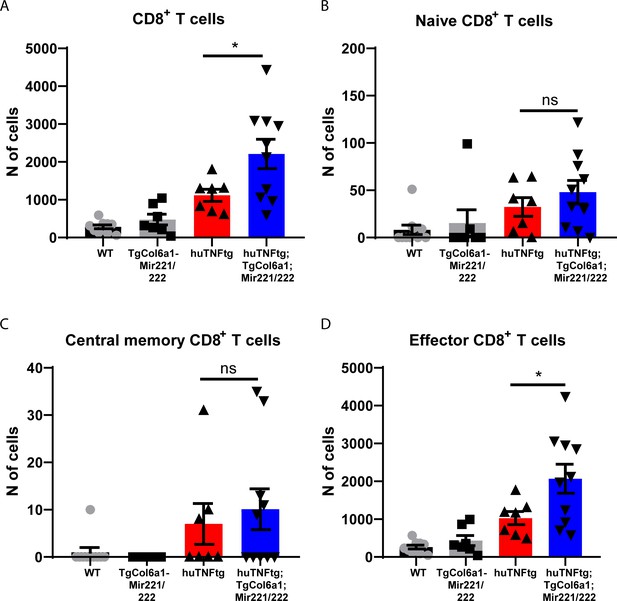
Mir221/222 overexpression leads to increased effector CD8+ T cell infiltration.
(A–D) Infiltration and characterization of CD8+ T cells in the ankle joints of 8-week-old WT (n = 10), TgCol6a1-Mir221/222 (n = 7), huTNFtg (n = 7), and huTNFtg;TgCol6a1-Mir221/222 mice (n = 10) quantified by FACS analysis (from three independent experiments). Student’s t-test statistical analysis was used, unpaired.
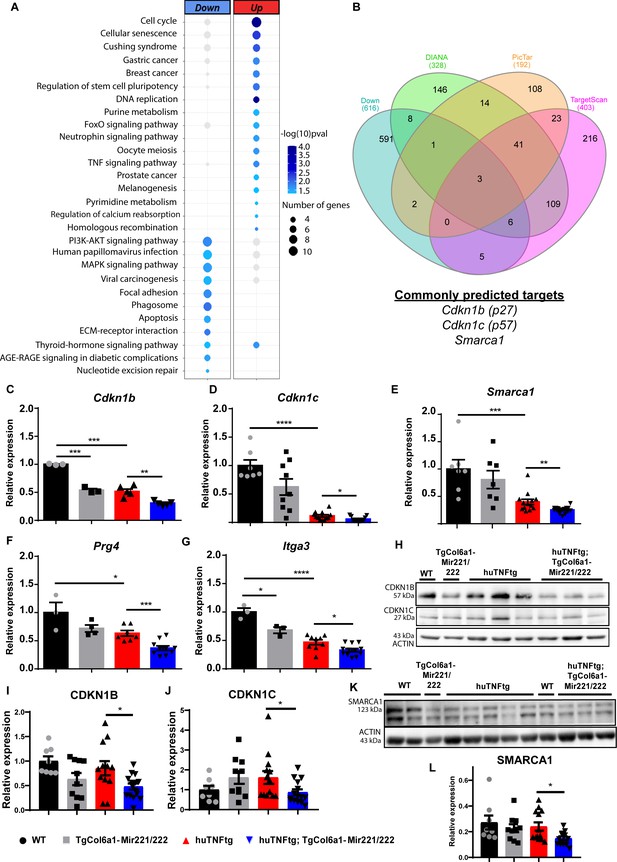
Mir221/222 regulate cell cycle signaling and ECM-related pathways in arthritis.
(A) Bubble plot of enriched KEGG pathways in the up- (red) and downregulated genes (blue) originated from the comparison of huTNFtg;TgCol6a1-Mir221/222 and huTNFtg bulk RNA-seq profiles. The color of the bubble signifies the statistical significance, while the size denotes the number of up-/downregulated genes found in the enriched term. (B) Venn diagram showing the overlap of genes predicted as Mir221/222 targets from DIANA, PicTar, and TargetScan and downregulated genes from huTNFtg compared to WT and huTNFtg;TgCol6a1-Mir221/222 compared to huTNFtg synovial fibroblasts (SFs). (C–G) Expression analysis as defined by qRT-PCR of Cdkn1b, Cdkn1c, Smarca1, Prg4, and Itga3 in cultured SFs from 8-week-old WT, TgCol6a1-Mir221/222, huTNFtg, and huTNFtg;TgCol6a1-Mir221/222 mice (n = 3–16). WT SFs were used as a reference. Data from 2 to 4 independent experiments. Student’s t-test statistical analysis was used, unpaired. In all experiments, B2m was used as a housekeeping gene for normalization. (H) Representative western blots depicting protein expression analysis of CDKN1B and CDKN1C in cultured SFs from 8-week-old WT, TgCol6a1-Mir221/222, huTNFtg, and huTNFtg;TgCol6a1-Mir221/222 mice. (I, J) Quantification of protein expression analysis as defined by western blots of CDKN1B and CDKN1C in cultured SFs from 8-week-old WT, TgCol6a1-Mir221/222, huTNFtg, and huTNFtg;TgCol6a1-Mir221/222 mice (n = 8–15). WT SFs were used as a reference. Data from four independent experiments. Student’s t-test statistical analysis was used, unpaired. In all experiments, ACTIN was used as a housekeeping gene for normalization. (K) Representative western blot depicting protein expression analysis of SMARCA1 in cultured SFs from 8-week-old WT, TgCol6a1-Mir221/222, huTNFtg, and huTNFtg;TgCol6a1-Mir221/222 mice. (L) Quantification of protein expression analysis as defined by western blots of SMARCA1 in cultured SFs from 8-week-old WT, TgCol6a1-Mir221/222, huTNFtg, and huTNFtg;TgCol6a1-Mir221/222 mice (n = 8–13). WT SFs were used as a reference. Data from four independent experiments. Student’s t-test statistical analysis was used, unpaired. In all experiments, ACTIN was used as a housekeeping gene for normalization. Data represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0,0001, ns = not significant.
-
Figure 4—source data 1
Differentially expressed genes as detected in bulk RNA sequencing originating from the comparison between different genotypes.
- https://cdn.elifesciences.org/articles/84698/elife-84698-fig4-data1-v1.zip
-
Figure 4—source data 2
List depicting downregulated genes stemming from the comparisons of huTNFtg compared to WT and huTNFtg;TgCol6a1-Mir221/222 compared to huTNFtg SFs along with Mir221/222 predicted targets using three different tools (DIANA-microT-CDS, Targetscan, and Pictar).
- https://cdn.elifesciences.org/articles/84698/elife-84698-fig4-data2-v1.zip
-
Figure 4—source data 3
Over-represented and down-represented KEGG pathways in the RNA expression profile of huTNFtg;TgCol6a1-Mir221/222 SFs compared to the huTNFtg.
- https://cdn.elifesciences.org/articles/84698/elife-84698-fig4-data3-v1.zip
-
Figure 4—source data 4
Uncropped blot of CDKN1B, CDKN1C, and ACTIN in cultured SFs from 8-week-old WT, TgCol6a1-Mir221/222, huTNFtg, and huTNFtg;TgCol6a1-Mir221/222 mice.
Exposure set at 1 s for CDKN1B and ACTIN.
- https://cdn.elifesciences.org/articles/84698/elife-84698-fig4-data4-v1.zip
-
Figure 4—source data 5
Uncropped blot of CDKN1B, CDKN1C, and ACTIN in cultured SFs from 8-week-old WT, TgCol6a1-Mir221/222, huTNFtg, and huTNFtg;TgCol6a1-Mir221/222 mice.
Exposure set at 7 s for CDKN1C.
- https://cdn.elifesciences.org/articles/84698/elife-84698-fig4-data5-v1.zip
-
Figure 4—source data 6
Uncropped blot of SMARCA1 and ACTIN in cultured SFs from 8-week-old WT, TgCol6a1-Mir221/222, huTNFtg, and huTNFtg;TgCol6a1-Mir221/222 mice.
Exposure set at 2 s for SMARCA1 and ACTIN.
- https://cdn.elifesciences.org/articles/84698/elife-84698-fig4-data6-v1.zip
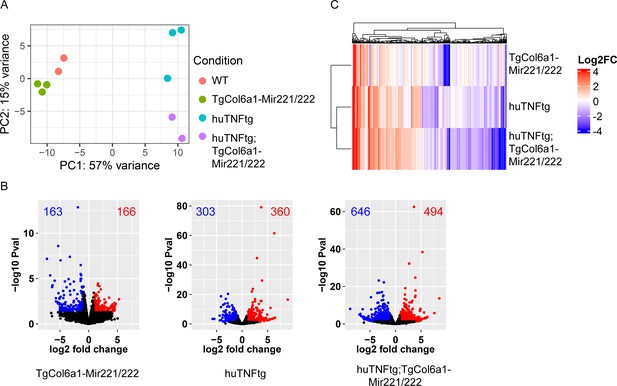
Comparisons between samples used for bulk RNA sequencing.
(A) Principal component analysis (PCA plot) of cultured synovial fibroblasts (SFs) from 8-week-old WT (n = 2), TgCol6a1-Mir221/222 (n = 3), huTNFtg (n = 3), and huTNFtg;TgCol6a1-Mir221/222 mice (n = 2). (B) Volcano plots of deregulated genes in SFs from TgCol6a1-Mir221/222, huTNFtg, and huTNFtg;TgCol6a1-Mir221/222 compared to WT SF expression profile. Nonsignificant deregulated genes are depicted with black color, significantly upregulated genes (p-value<0.05, log2FC > 1) with red, and significantly downregulated genes (p-value<0.05, log2FC < –1) with blue. (C) Heatmap showing log2 fold change values for the deregulated genes and contrasts plotted in (B). Hierarchical clustering has been performed for both genes and samples.
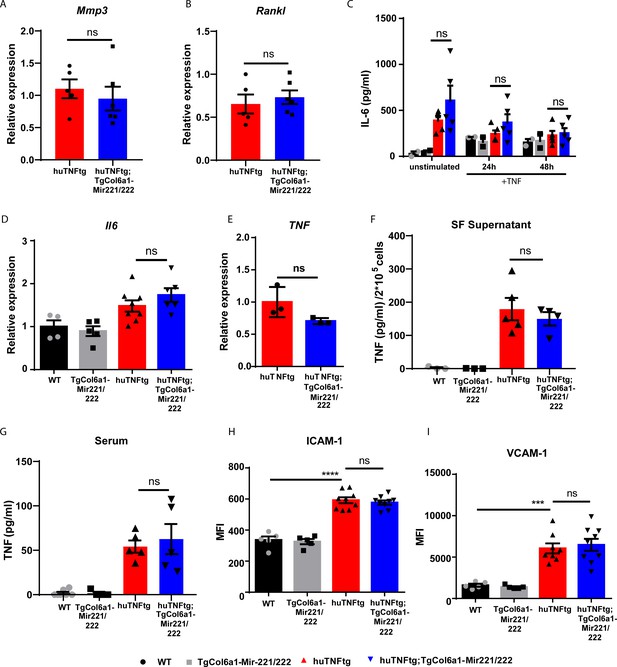
Mir221/222 do not regulate inflammatory signaling in synovial fibroblasts (SFs).
(A) qRT-PCR analysis of Mmp3 from huTNFtg (n = 5) and huTNFtg;TgCol6a1-Mir221/222 (n = 6) SFs. huTNFtg levels were used for normalization. B2m was used as a housekeeping gene. Student’s t-test statistical analysis was used, unpaired. (B) qRT-PCR analysis of Rankl from huTNFtg (n = 5) and huTNFtg;TgCol6a1-Mir221/222 (n = 6) SFs. huTNFtg levels were used for normalization. B2m was used as a housekeeping gene. Student’s t-test statistical analysis was used, unpaired. (C) IL-6 protein levels measured by ELISA in supernatants from WT (n = 2), TgCol6a1-Mir221/222 (n = 2), huTNFtg (n = 5), and huTNFtg;TgCol6a1-Mir221/222 mice (n = 5) SFs unstimulated or TNF stimulated for 24 or 48 hr. Student’s t-test statistical analysis was used, unpaired. (D) qRT-PCR analysis of Il6 from WT (n = 4), TgCol6a1-Mir221/222 (n = 5), huTNFtg (n = 8), and huTNFtg;TgCol6a1-Mir221/222 mice (n = 6) SFs. WT levels were used for normalization. B2m was used as a housekeeping gene. Student’s t-test statistical analysis was used, unpaired. (E) qRT-PCR analysis of TNF in SFs from huTNFtg (n = 3) and huTNFtg;TgCol6a1-Mir221/222 mice (n = 3). huTNFtg samples were used for normalization. B2m was used as a housekeeping gene. Student’s t-test statistical analysis was used, unpaired. (F, G) Human TNF protein levels measured by ELISA in SF supernatants and sera from WT (n = 3–7), TgCol6a1-Mir221/222 (n = 3–4), huTNFtg (n = 5), and huTNFtg;TgCol6a1-Mir221/222 mice (n = 4–5). Student’s t-test statistical analysis was used, unpaired. (H, I) Flow cytometry analysis of ICAM-1 and VCAM-1 expression as MFI in SFs from WT (n = 4), TgCol6a1-Mir221/222 (n = 5), huTNFtg (n = 8–9), and huTNFtg;TgCol6a1-Mir221/222 mice (n = 9). Student’s t-test statistical analysis was used, unpaired. Data represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0,0001, ns = not significant.
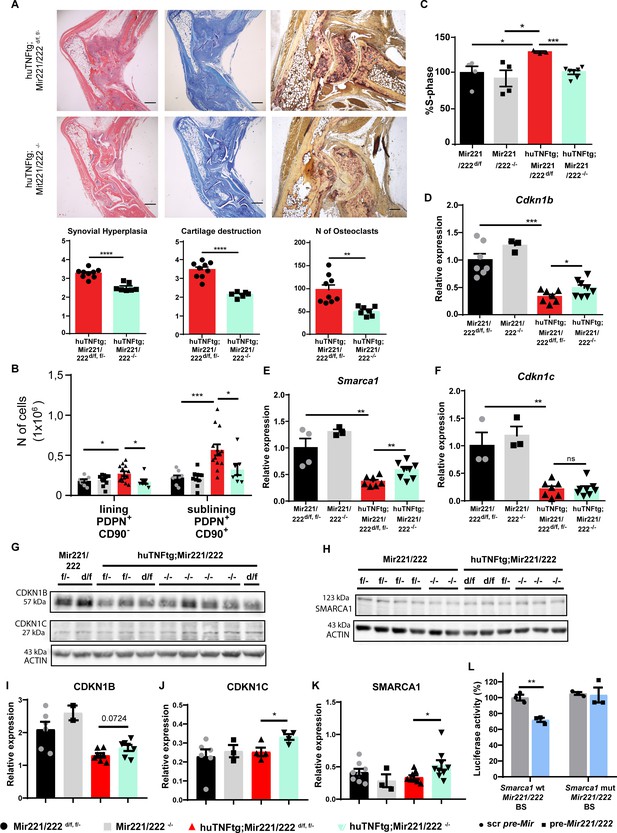
Deletion of Mir221/222 ameliorates arthritis in huTNFtg mice.
(A) Representative histological images of H&E, TB, and TRAP stained ankle joint sections and histological score of synovial hyperplasia, cartilage destruction, and osteoclast numbers from 9-week-old huTNFtg;Mir221/222 d/f, f/- (n = 9) and huTNFtg;Mir221/222 -/- mice (n = 6–7). Student’s t-test statistical analysis was used, unpaired. (B) Lining PDPN+CD90- and sublining PDPN+CD90+ synovial fibroblast (SF) number quantification in the ankle joints of 9-week-old Mir221/222 f/-, d/f (n = 8–9), Mir221/222 -/- (n = 10), huTNFtg;Mir221/222 f/-, d/f (n = 13) and huTNFtg;Mir221/222 -/- mice (n = 7) by FACS analysis (from three independent experiments). Student’s t-test statistical analysis was used, unpaired. (C) % fraction of cultured SFs that are in the S-phase of the cell cycle from 9-week-old Mir221/222 d/f (n = 4), Mir221/222 -/- (n = 4), huTNFtg;Mir221/222 d/f (n = 3), and huTNFtg;Mir221/222 -/- (n = 7) (from two independent experiments). Mir221/222 d/f SFs were used for normalization. Student’s t-test statistical analysis was used, unpaired. (D–F) Expression analysis as defined by qRT-PCR of Cdkn1b, Cdkn1c, and Smarca1 in cultured SFs from 9-week-old Mir221/222 f/-, d/f, Mir221/222 -/-, huTNFtg;Mir221/222 d/f and huTNFtg;Mir221/222 -/- (n = 3–8, from two independent experiments). Expression of Mir221/222 d/f, f/- SFs was used for normalization. Student’s t-test statistical analysis was used, unpaired. In all experiments, B2m was used as a housekeeping gene for normalization. (G, H) Representative western blots depicting protein expression analysis of CDKN1B, CDKN1C, and SMARCA1 in cultured SFs from 9-week-old Mir221/222 f/-, d/f, Mir221/222 -/-, huTNFtg;Mir221/222 d/f, and huTNFtg;Mir221/222 -/- mice. (I–K) Quantification of protein expression analysis as defined by western blots of CDKN1B, CDKN1C, and SMARCA1 in cultured SFs from 9-week-old Mir221/222 f/-, d/f, Mir221/222 -/-, huTNFtg;Mir221/222 d/f, and huTNFtg;Mir221/222 -/- mice (n = 2–9). WT SFs were used as a reference. Data from 2 to 3 independent experiments. Student’s t-test statistical analysis was used, unpaired. In all experiments, ACTIN was used as a housekeeping gene for normalization. (L) Luciferase activity in HEK293 cells transfected with constructs containing the wt Mir221/222 binding site (BS) of Smarca1 or the mutated one and co-transfected with either the control scramble pre-Mir or the pre-Mir221/222 for 72 hr. Scramble pre-Mir control co-transfected with the wt Smarca1 Mir221/222 BS served as a reference. Data from three independent experiments. Student’s t-test statistical analysis was used, unpaired. Scale bars: 600 μm and 300 μm. Data represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0,0001, ns = not significant.
-
Figure 5—source data 1
Uncropped blot of CDKN1B, CDKN1C, and ACTIN in cultured SFs from 9-week-old Mir221/222 f/-, d/f, huTNFtg;Mir221/222 d/f, and huTNFtg;Mir221/222-/- mice.
Exposure set at 1 s for CDKN1B.
- https://cdn.elifesciences.org/articles/84698/elife-84698-fig5-data1-v1.zip
-
Figure 5—source data 2
Uncropped blot of CDKN1B, CDKN1C, and ACTIN in cultured SFs from 9-week-old Mir221/222 f/-, d/f, huTNFtg;Mir221/222 d/f, and huTNFtg;Mir221/222-/- mice.
Exposure set at 2.4 s for ACTIN.
- https://cdn.elifesciences.org/articles/84698/elife-84698-fig5-data2-v1.zip
-
Figure 5—source data 3
Uncropped blot of CDKN1B, CDKN1C, and ACTIN in cultured SFs from 9-week-old Mir221/222 f/-, d/f, huTNFtg;Mir221/222 d/f, and huTNFtg;Mir221/222-/- mice.
Exposure set at 6.8 s for CDKN1C.
- https://cdn.elifesciences.org/articles/84698/elife-84698-fig5-data3-v1.zip
-
Figure 5—source data 4
Uncropped blot of SMARCA1 and ACTIN in cultured SFs from 9-week-old Mir221/222 f/-, d/f, Mir221/222-/-, huTNFtg;Mir221/222 d/f, and huTNFtg;Mir221/222-/- mice.
Exposure set at 3.8 s for SMARCA1 and ACTIN.
- https://cdn.elifesciences.org/articles/84698/elife-84698-fig5-data4-v1.zip
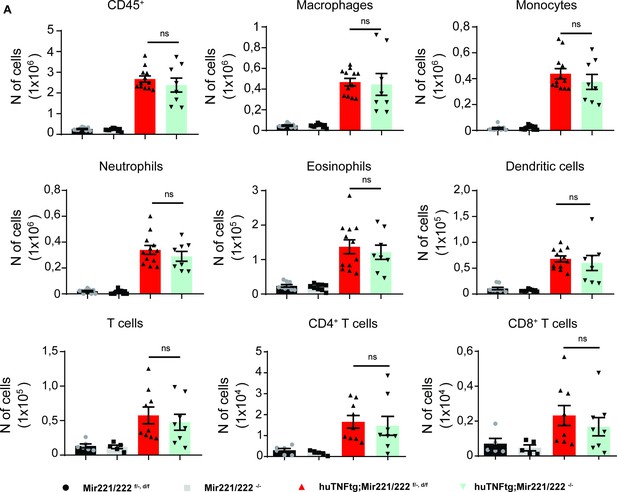
Mir221/222 deletion in experimental arthritis does not regulate inflammatory influx in the joints.
(A) Infiltration of CD45+ cells, macrophages, monocytes, neutrophils, eosinophils, dendritic cells, CD4+ T cells, and CD8+ T cells in the ankle joints of 9-week-old Mir221/222 f/-, d/f (n = 6–9), Mir221/222 -/- (n = 5–10), huTNFtg;Mir221/222 f/-, d/f (n = 9–12) huTNFtg;Mir221/222 -/- mice (n = 8–10) by FACS analysis (from three independent experiments). Student’s t-test statistical analysis was used, unpaired. Data represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0,0001, ns = not significant.
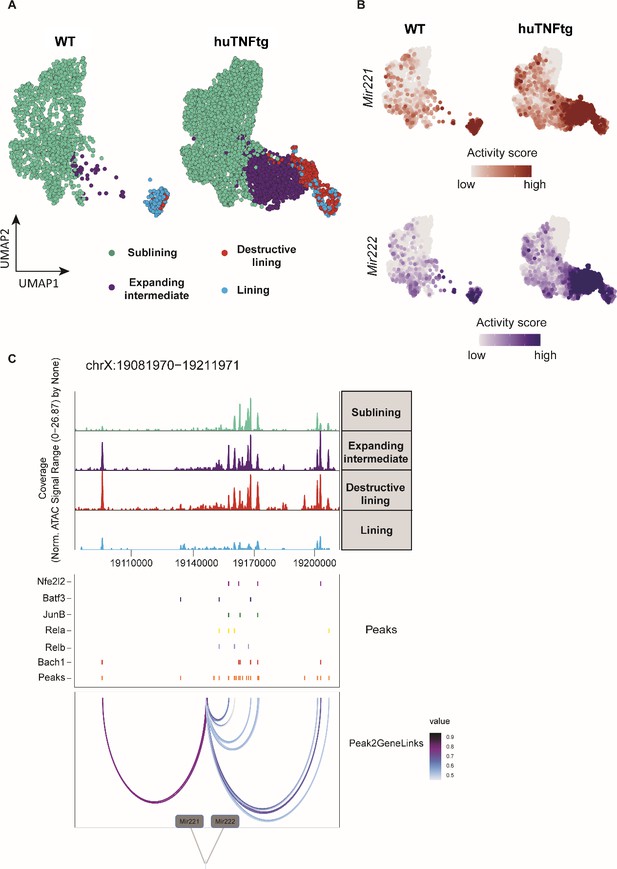
Mir221/222 gene activity is increased in the pathogenic subclusters of the expanding intermediate and lining compartment.
(A) UMAP projection of 6046 synovial fibroblast nuclei obtained by scATAC-seq from WT and huTNFtg samples. Cells are grouped in four categories: sublining (light green), expanding intermediate (purple), lining (light blue), and destructive lining (red). (B) Feature plots, in UMAP space, depicting gene activity scores of Mir221 (red) and Mir222 (blue) in WT and huTNFtg samples. (C) Genome accessibility track visualization of the extended regulatory space of Mir221 and Mir222 (chrX:19,081,970–19,211,971), with TF binding site and peak-to-gene linkage information, in huTNFtg samples. Upper: the genome track shows increased accessibility in expanding intermediate and destructive lining clusters. Middle: all reproducible peaks are shown, coupled with annotated CISBP binding information for Bach1, Rela, Relb, Nfe2l2, Batf3, and Junb TFs. Lower: putative regulatory linkages between Mir221-Mir222 genes and reproducible peaks are illustrated. Links between genes and peaks are colored by correlation (Pearson coefficient) of peak accessibility and gene activity scores.
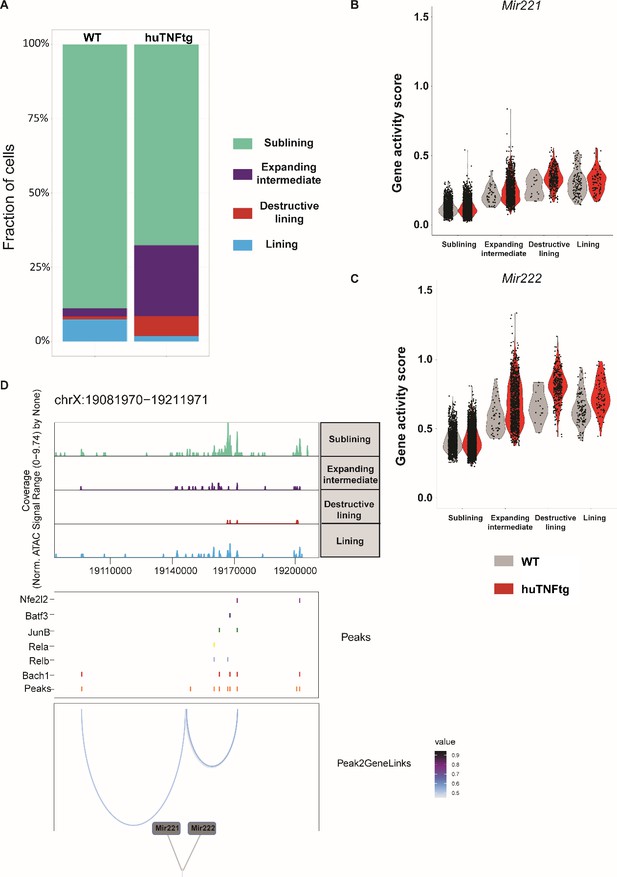
Mir221/222 gene activity in synovial fibroblast (SF) clusters in normal and arthritic state.
(A) Stacked barchart showing relative abundances (%) of cell clusters (as described in Figure 6A) across WT and huTNFtg samples. Both intermediate and destructive lining populations are expanding in disease state. (B, C) Violin plots exhibiting gene activity scores for Mir221 and Mir222 genes. Split view is selected in order to highlight the differences between WT (shown in gray) and huTNFtg (shown in red) samples across the different clusters. (D) Genome accessibility track visualization of the extended regulatory space of Mir221 and Mir222 (chrX:19,081,970–19,211,971), with TF binding site and peak-to-gene linkage information, in WT sample. Upper: the genome track shows increased accessibility in intermediate and destructive lining clusters. Middle: all reproducible peaks are shown, coupled with annotated CISBP binding information for Bach1, Rela, Relb, Junb, Batf3, and Nfe2l2 TFs. Lower: putative regulatory linkages between Mir221-Mir222 genes and reproducible peaks are illustrated. Links between genes and peaks are colored by correlation (Pearson coefficient) of peak accessibility and gene activity scores.
Additional files
-
Supplementary file 1
Primers used for mouse genotyping and qPCR primer sequences (5′–3′) are provided in (a and b).
- https://cdn.elifesciences.org/articles/84698/elife-84698-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/84698/elife-84698-mdarchecklist1-v1.pdf