Pupil size reflects activation of subcortical ascending arousal system nuclei during rest
Figures

Whole-brain pupil-blood oxygen level-dependent (pupil-BOLD) coupling in comparison to previous studies.
Neural correlates of pupil size from the analysis using the convolution approach (A) reproduced from Figure 1B of Schneider et al., 2016 and (B) reproduced from Figure 2A of Murphy et al., 2014. Note that we only refer to the red and yellow activation in the figure from Murphy et al., 2014. Statistical parametric maps are thresholded at p<0.001, uncorrected, for visualization purposes only. Whole-brain cluster-level family-wise error (FWE)-corrected inferential statistics, in MNI space, are reported in Tables 1 and 2 (n = 70). Statistical maps used to generate this figure is available in Figure 1—source data 1.
© 2016, Elsevier Inc. Whole-brain pupil-BOLD coupling in comparison to previous studies. Neural correlates of pupil size from the analysis using the convolution approach from (A) reproduced from Figure 1B of Schneider et al., 2016, with permission of Elsevier Inc. and (B) reproduced from Figure 2A of Murphy et al., 2014, with permission of Wiley Periodicals Inc. These panels are not covered by the CC-BY 4.0 license and further reproduction would need permission from the copyright holders.
-
Figure 1—source data 1
Statistical maps used in Figure 1, Tables 1 and 2.
- https://cdn.elifesciences.org/articles/84822/elife-84822-fig1-data1-v1.zip
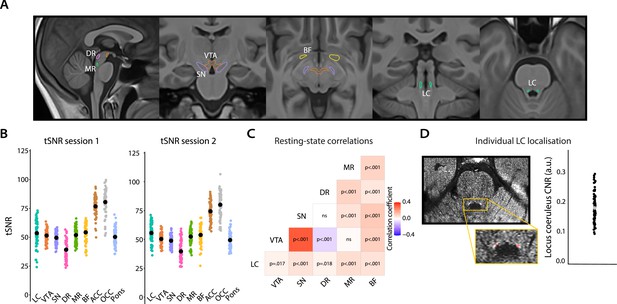
Overview of ROI definition and validation of the quality of subcortical fMRI data.
(A) All subcortical ROIs overlaid on the group T1 template. (B) Individual data points showing the temporal signal-to-noise ratio for each ROI for session 1 (left) and session 2 (right; black points indicate the mean). (C) Correlation matrix showing that activity in subcortical nuclei co-varied positively with activity in other subcortical nuclei, with the strongest coupling present between the VTA and SN, which is to be expected given their close proximity, and the weakest (negative correlation) between the VTA and DR. Note: correlations were FDR-corrected and controlled for activity in the pons (n = 70). (D) FSE image of an example participant. Hyperintensities corresponding to the LC are visible in the yellow box (top). Using the FSE images, the LC (red) was manually delineated on the individual level following established protocols (Clewett et al., 2016; Mather et al., 2017). The graph shows the LC contrast-to-noise ratio for all participants (n = 67). The gray dot indicates the grand mean. LC, locus coeruleus; VTA, ventral tegmental area; SN, substantia nigra; DR, dorsal raphe; MR, medial raphe; BF, basal forebrain; ACC, anterior cingulate cortex; OCC, calcarine sulcus; CNR, contrast-to-noise ratio; ROI, region of interest; fMRI, functional magnetic resonance imaging; FDR, false discovery rate. Source data used to generate this figure is available in Figure 2—source data 1.
-
Figure 2—source data 1
These csv files contains the data for Figure 2B–D.
- https://cdn.elifesciences.org/articles/84822/elife-84822-fig2-data1-v1.zip
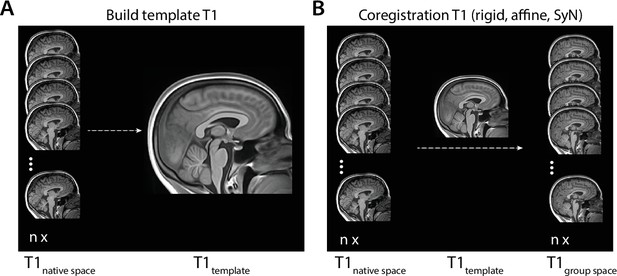
Overview of template building and coregistration steps performed in ANTs.
(A) Individual T1 images (T1native space) were submitted to a template building algorithm where they were aligned and used to generate a group average (T1template). (B) Individual T1 images (T1native space) were coregistered to the T1 template in a separate coregistration step, generating individual T1 images in group space (T1group space). SyN, symmetric normalization.
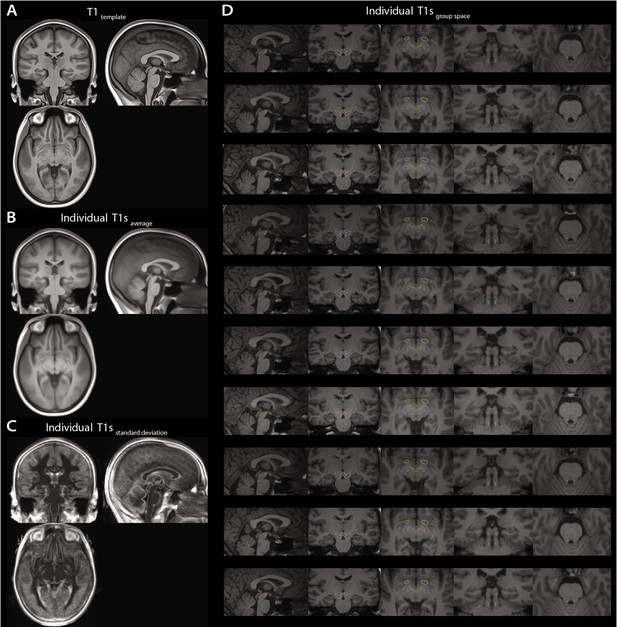
Anatomical verification of accurate registration.
(A) The sample-specific template T1. (B) The average of all individual warped T1 images. (C) The standard deviation of all individual warped T1 images. (D) Individual examples displaying the ascending arousal system (AAS) region of interest (ROI) masks on top of the individual warped T1.

Overview of the analysis using region- and participant-specific estimates of the HRF.
(A) One participant’s pre-processed LC BOLD signal (concatenated across the two sessions), evaluated against a chosen threshold (>1 SD) to extract onsets of spontaneous neural events (indicated in yellow). (B) Estimated HRFs for each participant and ROI. Black lines indicate the average across participants. (C) The number of spontaneous neural events that were used to estimate the region- and participant-specific HRFs. Black dots indicate the mean. (D) TTP of the HRF for each participant and ROI. Black dots indicate the average across participants. Overview of the pupil data processing pipeline of one randomly chosen participant for pupil size (E) and the pupil derivative (F). The plots portray the raw pupil data (blue), the pupil time series down-sampled to the TR (red), and the convolved pupil time series (for LC HRF, green). The extracted t-values from the first-level resting-state HRF analysis for pupil size (G) and the pupil derivative (H). p-Values refer to one-sample t-tests (difference from zero; FDR-corrected; n = 70). LC, locus coeruleus; VTA, ventral tegmental area; SN, substantia nigra; DR, dorsal raphe; MR, median raphe; BF, basal forebrain; ACC, anterior cingulate cortex; OCC, calcarine sulcus; HRF, hemodynamic response function; BOLD, blood oxygen level-dependent; TTP, time-to-peak; FDR, false discovery rate. Source data used to generate this figure available in Figure 3—source data 1.
-
Figure 3—source data 1
These csv tables contain the data used for Figure 3B–D, G–H.
- https://cdn.elifesciences.org/articles/84822/elife-84822-fig3-data1-v1.zip
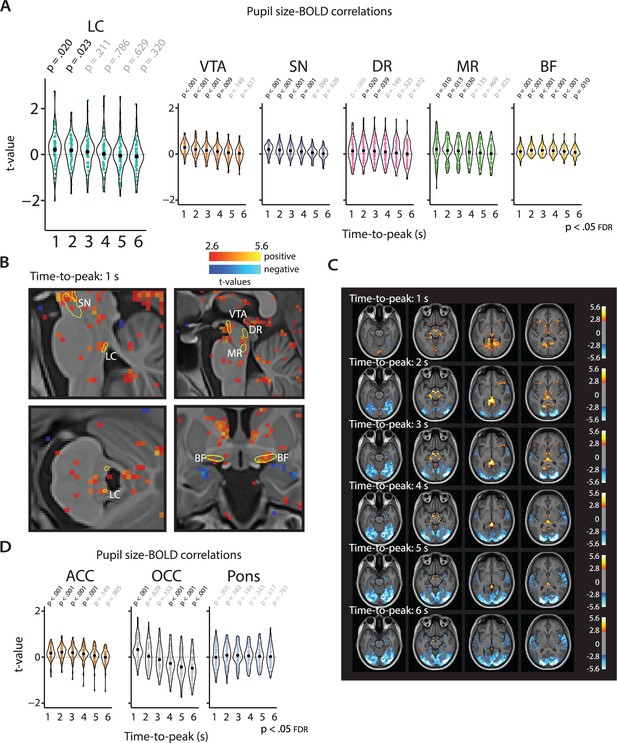
Pupil-BOLD correlations based on systematic adjustment of the TTP of the HRF.
Graphs show the extracted t-statistics as a function of the systematically adjusted TTP for each participant in the subcortical ROIs (A) and the validation and control ROIs (D). p-Values refer to one-sample t-tests (difference from zero; FDR-corrected). Black dots indicate the mean. (B) Statistical maps showing pupil-BOLD correlations in the subcortex for the 1 s TTP. (c) Statistical maps showing unsmoothed pupil-BOLD correlations across the cortex for each TTP (1–6 s). All statistical maps were thresholded at p<0.005 (uncorrected) for visualization purposes only (n = 70). LC, locus coeruleus; VTA, ventral tegmental area; SN, substantia nigra; DR, dorsal raphe; MR, median raphe; BF, basal forebrain; ACC, anterior cingulate cortex; OCC, calcarine sulcus; BOLD, blood oxygen level-dependent; FDR, false discovery rate; TTP, time-to-peak; HRF, hemodynamic response function; ROI, region of interest. Source data used to generate this figure is available in Figure 4—source data 1.
-
Figure 4—source data 1
This folder contains the csv files and statistical maps used to make Figure 4.
- https://cdn.elifesciences.org/articles/84822/elife-84822-fig4-data1-v1.zip
-
Figure 4—source data 2
This contains the csv tables used in Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/84822/elife-84822-fig4-data2-v1.zip
-
Figure 4—source data 3
This folder contains the csv tables and statistical maps used in Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/84822/elife-84822-fig4-data3-v1.zip
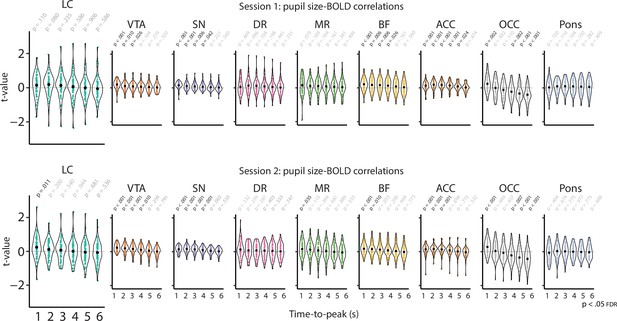
Pupil size-BOLD correlations based on systematic adjustment of the TTP of the HRF for both sessions separately.
Graphs show the individual t-statistics (black point indicates the mean) of the pupil size-BOLD signal correlation as a function of the systematically adjusted TTP for all ROIs, p-Values refer to one-sample t-tests (difference from zero; FDR-corrected; n = 70). These results show that smaller ROIs, in particular the LC, DR, and MR, are more prone to a drop in statistical power when the data is halved. LC, locus coeruleus; VTA, ventral tegmental area; SN, substantia nigra; DR, dorsal raphe; MR, medial raphe; ACC, anterior cingulate cortex; OCC, calcarine sulcus; BOLD, blood oxygen level-dependent; FDR, false discovery rate; TTP, time-to-peak; HRF, hemodynamic response function; ROI, region of interest. Source data used to generate this figure is available in Figure 4—source data 2.
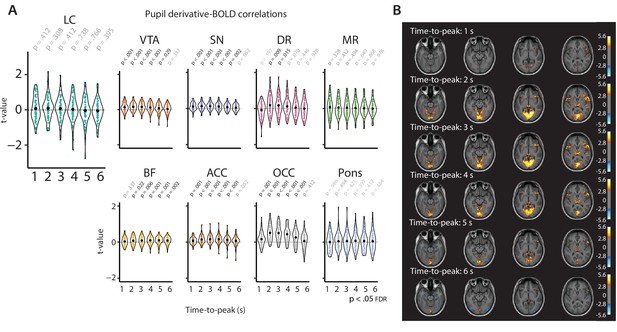
Pupil derivative-BOLD correlations based on systematic adjustment of the TTP of the HRF.
(A) Graphs show the individual t-statistics (black point indicates the mean) of the pupil derivative-BOLD signal correlation as a function of the systematically adjusted TTP for all ROIs, p-Values refer to one-sample t-tests (difference from zero; FDR-corrected). (B) The corresponding whole-brain associations between BOLD signal and the pupil derivative as a function of the TTP of the HRF (1 s TTP: top panel to 6 s TTP: bottom panel). The warm colors reflect areas in which BOLD activity is positively correlated with pupil derivative. The cold colors reflect areas where BOLD activity is negatively correlated with the pupil derivative. Both positive and negative contrast maps were sampled at p<0.005 (uncorrected) for visualization only (n = 70). LC, locus coeruleus; VTA, ventral tegmental area; SN, substantia nigra; DR, dorsal raphe; MR, medial raphe; ACC, anterior cingulate cortex; OCC, calcarine sulcus; BOLD, blood oxygen level-dependent; FDR, false discovery rate; TTP, time-to-peak; HRF, hemodynamic response function; ROI, region of interest. Source data used to generate this figure is available in Figure 4—source data 3.
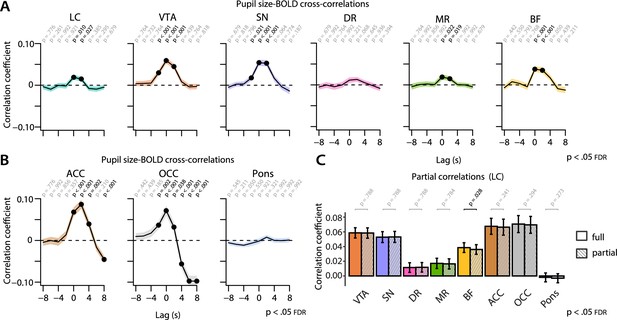
Cross-correlations between the unconvolved pupil time series and the BOLD time series at various time lags.
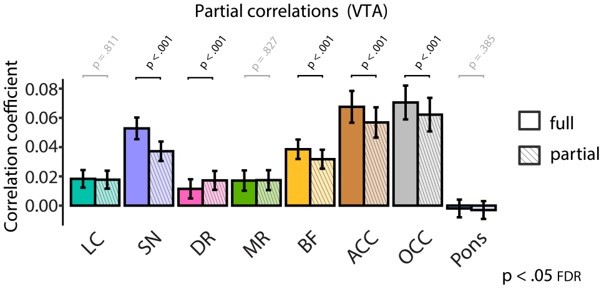
Pupil-BOLD time-series cross-correlations for all AAS nuclei ROIs (A) and control ROIs (B). Negative (positive) time lags indicate that the pupil signal precedes (follows) the BOLD signal. p-Values refer to one-sample t-tests for the corresponding time bin. (C) Statistical comparisons of pupil size-BOLD signal coupling at lag = 0 s before (full) and after regressing out LC BOLD signal (partial). Statistics refer to two-tailed t-tests (FDR-corrected) comparing the full and partial correlation coefficients. Black lines (A, B) and bar plots (C) indicate the grand mean and shaded regions (A, B) and error bars (C) indicate ± standard error of the mean (SEM). Black font and black dots indicate significant time bins (p<0.05, FDR-corrected) (n = 70). BOLD, blood oxygen level-dependent; AAS,ascending arousal system; ROI, region of interest; LC,locus coeruleus; FDR, false discovery rate. Source data used to generate this figure is available in Figure 5—source data 1.
-
Figure 5—source data 1
These csv tables contain the data used for Figure 5.
- https://cdn.elifesciences.org/articles/84822/elife-84822-fig5-data1-v1.zip
-
Figure 5—source data 2
This contains the csv files used in Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/84822/elife-84822-fig5-data2-v1.zip
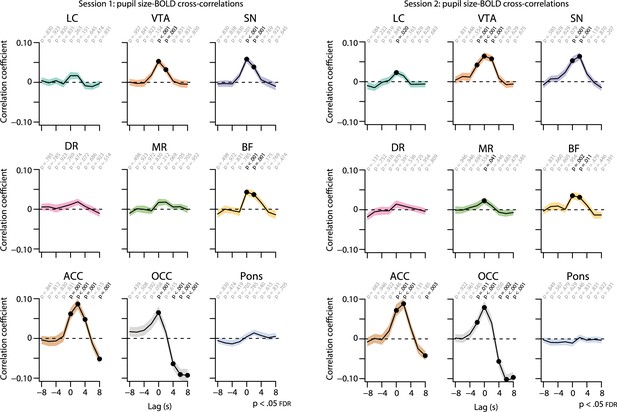
Cross-correlations between the unconvolved pupil size time series and the BOLD time series at various time lags for each resting-state session.
The BOLD time series was shifted forward and backward by 8 s in steps of 2 s. Positive (negative) time lags indicate that the pupil signal precedes (follows) the BOLD signal. Black lines indicate the grand mean, and shaded regions indicate the standard error of the mean. Black dots indicate significant one-sample t-tests (p<0.05, FDR-corrected; n = 70). LC, locus coeruleus; VTA, ventral tegmental area; SN, substantia nigra; DR, dorsal raphe; MR, medial raphe; ACC, anterior cingulate cortex; OCC, calcarine sulcus; BOLD, blood oxygen level-dependent; FDR. false discovery rate. Source data used to generate this figure available in Figure 5—source data 1.
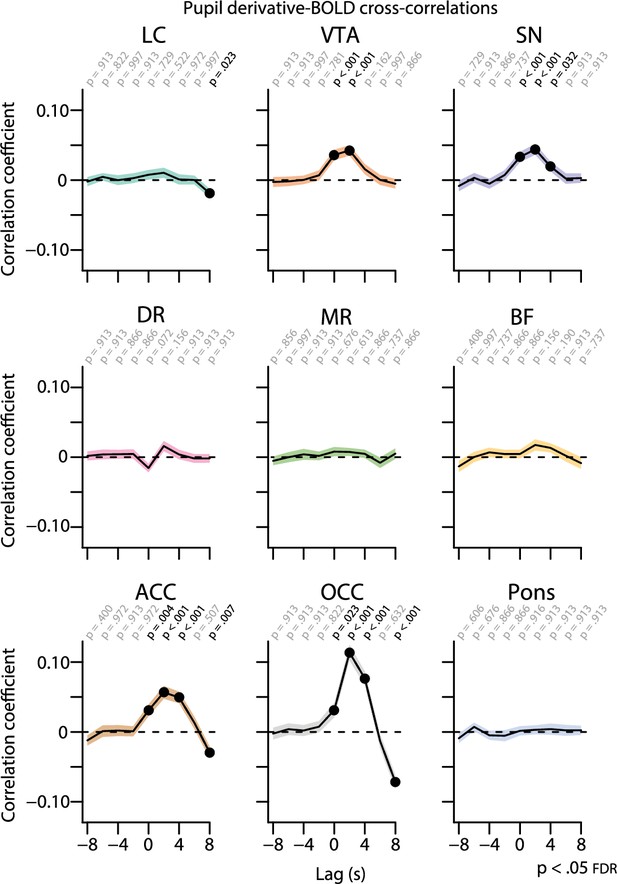
Cross-correlations between the unconvolved pupil derivative time series and the BOLD time series at various time lags.
The BOLD time series was shifted forward and backward by 8 s in steps of 2 s. Positive (negative) time lags indicate that the pupil signal precedes (follows) the BOLD signal. Shaded regions indicate the standard error of the mean. Black dots indicate significant one-sample t-tests (p<0.05, FDR-corrected; n = 70). LC, locus coeruleus; VTA, ventral tegmental area; SN, substantia nigra; DR, dorsal raphe; MR, medial raphe; ACC, anterior cingulate cortex; OCC, calcarine sulcus; BOLD, blood oxygen level-dependent; FDR, false discovery rate. Source data used to generate this figure is available in Figure 5—source data 2.
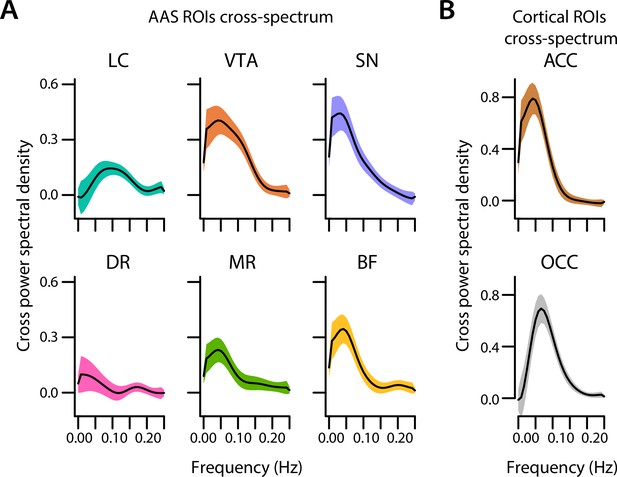
Cross-spectral power density analysis.
Cross-spectral power density for subcortical ROIs (A) and cortical ROIs (B) averaged across participants. Black lines indicate the grand mean and shaded regions indicate ± SEM (n = 70). LC, locus coeruleus; VTA, ventral tegmental area; SN, substantia nigra; DR, dorsal raphe; MR, median raphe; BF, basal forebrain; ACC, anterior cingulate cortex; OCC, calcarine sulcus; ROI, region of interest. Source data used to generate this figure is available in Figure 6—source data 1.
-
Figure 6—source data 1
This folder contains the csv tables used in Figure 6.
- https://cdn.elifesciences.org/articles/84822/elife-84822-fig6-data1-v1.zip
T1template -> T1MNI-space registration check.
Template image warped into MNI-space (yellow outline) overlaid on top of the original MNI-space T1.
Tables
Regions showing pupil-BOLD coupling using the convolution methods from Schneider et al., 2016.
| Cluster | ||
|---|---|---|
| K | pcorr | |
| Positive | ||
| Thalamus (R), posterior cingulate cortex (R/L) | 871 | <0.001 |
| Rectus (L) | 199 | 0.010 |
| Cerebellum (R) | 142 | 0.043 |
| Cerebellum crus (L) | 250 | 0.003 |
| Negative | ||
| Cerebellum crus (R/L), cerebellum 6 (R/L), cerebellum 4/5 (R/L), lingual gyrus (R/L), calcarine (R/L), fusiform gyrus (R/L), cuneus (R/L), precuneus (R/L), cerebellum 4+5 (L), cerebellar vermis, hippocampus (R/L), parahippocampal gyrus (R), amygdala (R/L), thalamus (R), superior occipital gyrus (R/L), middle occipital gyrus (R/L), inferior occipital gyrus (R/L), superior parietal gyrus (L), inferior temporal gyrus (R/L), middle temporal gyrus (R/L), superior temporal gyrus (R/L), insula (R), postcentral gyrus (L), precentral gyrus (L), paracentral lobule (R/L), supplementary motor area (R/L), middle cingulate gyrus | 65223 | <0.001 |
-
Reported clusters survived whole-brain family-wise error (FWE) correction at the cluster level (pFWE=0.05). Source data for Table 1 available in Figure 1—source data 1.
-
R, right; L, left; pcorr, whole-brain-corrected cluster p-values; k, cluster size; BOLD, blood oxygen level-dependent.
Regions showing pupil-BOLD coupling using convolution methods from Murphy et al., 2014.
| Cluster | ||
|---|---|---|
| K | pcorr | |
| Middle occipital gyrus (R/L), superior occipital gyrus (R/L), calcarine gyrus (R/L), cuneus (R/L), precuneus (R/L), angular gyrus (R/L), fusiform gyrus (R/L), cerebellum (R/L), middle temporal pole (R/L), inferior temporal pole (L), insula (R), inferior parietal lobule (R/L), superior parietal lobule (L), postcentral gyrus (R/L), middle frontal gyrus (R/L), medial frontal gyrus (R/L), inferior frontal gyrus (R/L), superior frontal gyrus (R/L), posterior cingulate gyrus, middle cingulate gyrus, anterior cingulate gyrus, supplementary motor area (R/L), middle frontal orbital (R/L), inferior frontal orbital (R/L), cerebellum crus II (R/L), cerebellum crus I (R/L), cerebellum 8 (R/L), cerebellum 9 (R/L), Pons | 91471 | <0.001 |
| Rectus (R/L) | 146 | 0.010 |
-
Reported clusters survived whole-brain family-wise error (FWE) correction at the cluster level (pFWE=0.05). Source data for Table 2 is available in Figure 1—source data 1.
-
R, right; L, left; pcorr, whole-brain-corrected cluster p-values; k, cluster size;.BOLD, blood oxygen level-dependent.