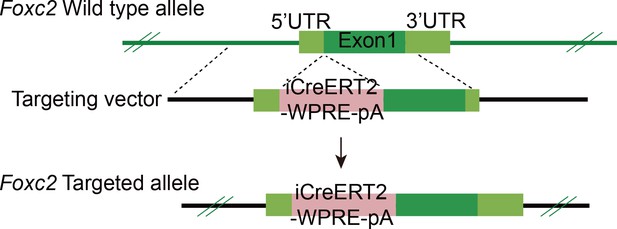
Identification of quiescent FOXC2+ spermatogonial stem cells in adult mammals
Figures
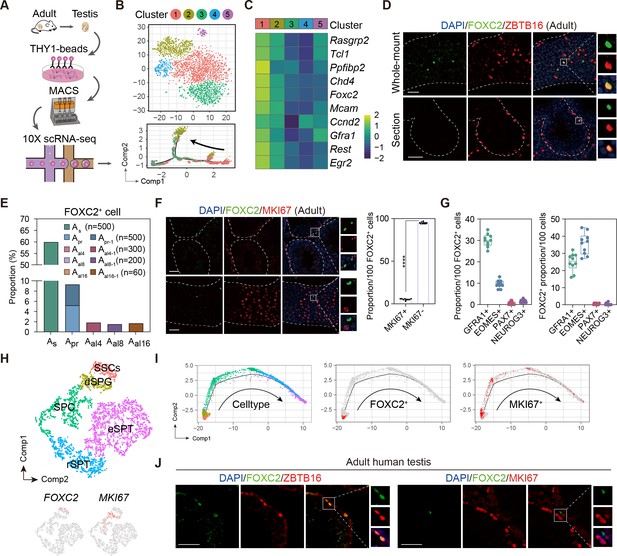
Identification of the FOXC2+ spermatogonial stem cells (SSCs) in adult mouse and human testis.
(A) Schematic illustration of the single-cell analysis workflow. (B) t-SNE plot and developmental trajectory of all undifferentiated spermatogonia (uSPG), colored by cluster. (C) Heatmap of the top 10 differentially expressed genes (DEGs) in Cluster1. (D) Immunostaining for ZBTB16 (red), FOXC2 (green), and DAPI (blue) in testicular paraffin sections from wild-type adult C57 mice. Scale bar, 50 μm; C57, C57BL/6J. (E) The proportion of FOXC2+ cells in different uSPG subtypes. (F) Immunostainings for MKI67 (red), FOXC2 (green), and DAPI (blue) in adult mice testis and the proportion of MKI67+ cells in FOXC2+ population (n=10). Scale bar, 50 μm; values, mean ± s.e.m.; p-values were obtained using two-tailed t-tests (****p-value <0.0001). (G) The co-expression proportion between the FOXC2 and differential known SSCs makers (n=10). (H) t-SNE plot of germ cells in adult human testis (GSE112013), colored by germ cell type. Feature plot showing the expression patterns of FOXC2 and MKI67 in human germ cells. (I) The developmental trajectory of the human germ cells, colored by germ cell type, FOXC2 expression cells (red), or MKI67 expression cells (red). (J) Immunostaining for ZBTB16/MKI67 (red), FOXC2 (green), and DAPI (blue) in testicular paraffin sections from adult humans.
-
Figure 1—source data 1
Excel spreadsheet with the list of the top 30 differentially expressed genes of different clusters.
- https://cdn.elifesciences.org/articles/85380/elife-85380-fig1-data1-v1.xlsx
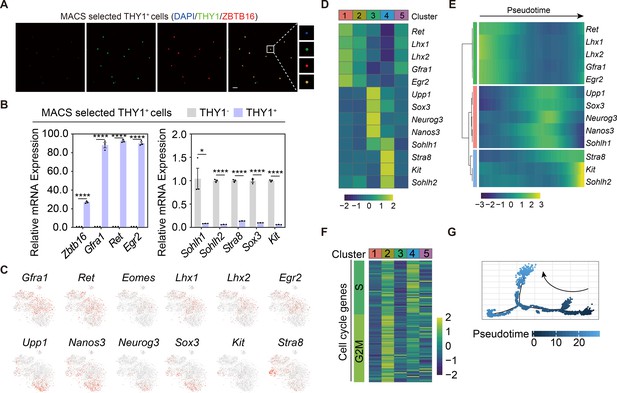
Validation and characterization of the magnetic-activated cell sorting (MACS)-sorted THY1+ undifferentiated spermatogonia (uSPG) from wild-type adult C57 mice.
(A) Immunostainings of DAPI (blue), THY1 (green), and ZBTB16 (red) in the MACS-sorted THY1+ cells (n=5). Scale bar, 50 μm. (B) Quantitative RT-PCR analysis of uSPG and differentiating spermatogonia (dSPG) markers expressed in the MACS-sorted THY1+ cells (n=3). Values, mean ± s.e.m.; p-values were obtained using two-tailed t-tests (ns >0.05, *p-value <0.05, **p-value <0.01, ***p-value <0.001, ****p-value <0.0001). (C) Feature plots showing the expression pattern of classic SPG markers (stemness and differentiation). (D) Heatmap showing the expression pattern of markers for SPG in different clusters. (E) Expression pattern dynamics of the SPG markers with pseudotime progression. (F) Heatmap showing the expression pattern of markers for cell cycle phase in different clusters. (G) The developmental trajectory of the overall SPG, colored by pseudotime.
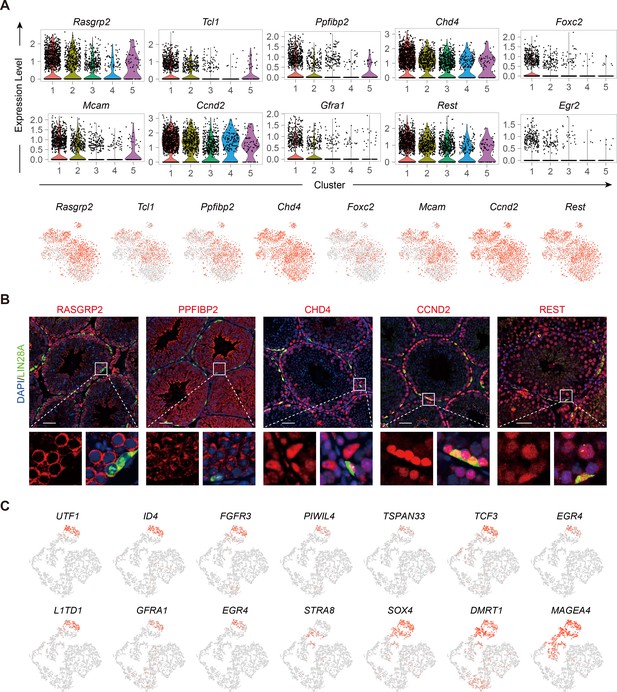
Expression of top 10 differentially expressed genes (DEGs) of Cluster1 in Figure 1B and classic spermatogonial stem cell (SSC) and SPG markers in adult human germ cells.
(A) Feature plots and violin plots of the top 10 DEGs of Cluster1. (B) Immunostainings for LIN28A (red), DAPI (blue), and newly found markers (green) in testicular paraffin sections from adult mice. Scale bar, 50 μm. (C) Feature plots showing the expression pattern of classic SSCs and SPG markers in adult human germ cells.
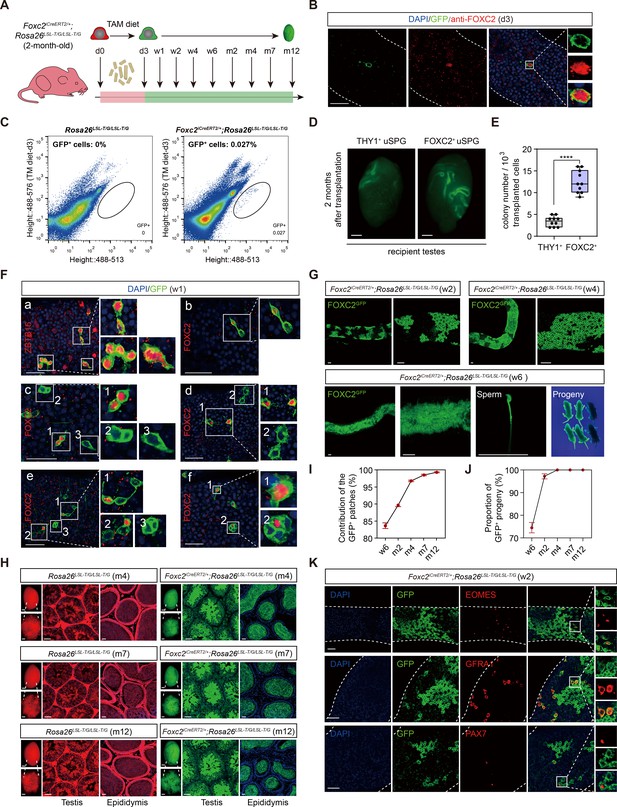
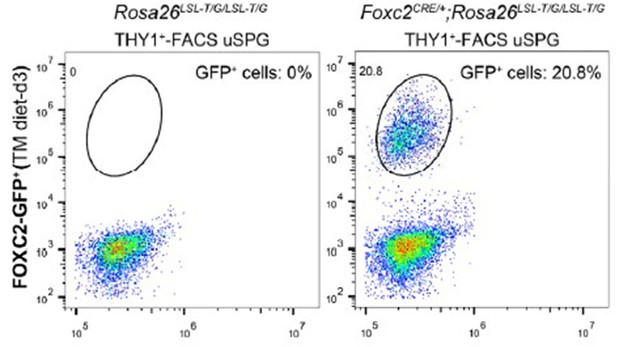
Lineage tracing and functional validation of FOXC2+ spermatogonial stem cells (SSCs) in Foxc2iCreERT2/+;Rosa26LSL-T/G/LSL-T/G mice.
(A) Schematic illustration of the lineage tracing workflow for FOXC2+ cells. (B) Immunostainings for DAPI (blue) and FOXC2 (red) at day 3 post TAM induction. Scale bar, 50 μm; d, day. (C) Fluorescence-activated cell sorting (FACS) analysis of GFP+ populations derived from Rosa26LSL-T/G/LSL-T/G or Foxc2iCreERT2/+;Rosa26LSL-T/G/LSL-T/G mice at day 3 post TAM induction. (D, E) The recipient mice testes (D) and colony numbers (E) 2 months after transplantation (n=10) of the FACS-sorted GFP+ cells from the Foxc2iCreERT2/+;Rosa26LSL-T/G/LSL-T/G mice 3 days after TAM diet and the FACS-sorted THY1+ cells from adult mice. Scale bar, 1 mm; values, mean ± s.e.m.; p-values were obtained using two-tailed t-tests (****p-value <0.0001). (F) Immunostaining for DAPI (blue), ZBTB16/FOXC2 (red), and GFP (green) at week 1 post TAM induction (scale bar, 50 μm). (G) Seminiferous tubules of Foxc2iCreERT2/+;Rosa26LSL-T/G/LSL-T/G mice 2, 4, and 6 weeks post TAM induction. Scale bar, 50 μm. (H) Testes (scale bar, 1 mm), seminiferous tubules, and epididymis (scale bar, 50 μm) at months 4, 7, and 12 post TAM induction in Foxc2iCreERT2/+;Rosa26LSL-T/G/LSL-T/G mice. (I, J) The GFP+ patches (I) and progeny (J) population dynamics (n=10). Values, mean ± s.e.m. (K) Immunostainings for DAPI (blue), EOMES (red), GFRA1 (red), or PAX7 (red) in GFP+ population at week 2 post TAM induction. Scale bar, 50 μm.
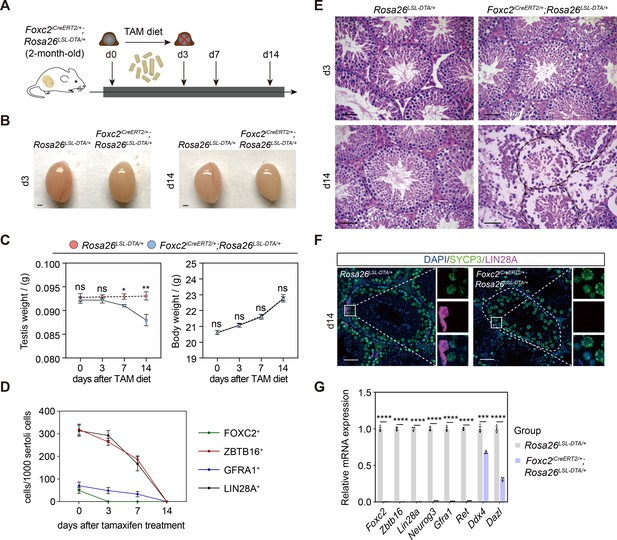
Specific ablation of FOXC2+ spermatogonial stem cells (SSCs) and phenotypic validation in Foxc2iCreERT2/+;Rosa26LSL-DTA/+ mice.
(A) Schematic illustration of the lineage tracing workflow for FOXC2+ cells. (B–D) Phenotypic validation of the Rosa26LSL-DTA/+ and Foxc2iCreERT2/+;Rosa26LSL-DTA/+ mice (n=5) for testes size (B), testis weight and body weight (C), and hematoxylin and eosin (HE) staining of the testes (D). Scale bars in (B), 1 mm; in (D), 50 μm; d, day; values were mean ± s.e.m.; p-values were obtained using two-tailed t-tests (ns >0.05, *p-value <0.05, **p-value <0.01). (E) ZBTB16+, GFRA1+, LIN28A+, and FOXC2+ SPG populations dynamics. Values, mean ± s.e.m. (n=10); p-values were obtained using one-way ANOVA followed by Tukey test (ns >0.05, *p-value <0.05, **p-value <0.01, ****p-value <0.0001). (F) Immunostainings for DAPI (blue), SYCP3 (green), and LIN28A (magenta) at day 14 post TAM induction. d, day; scale bar, 50 μm. (G) Quantitative RT-PCR analysis of SPG markers expression in the testes of the Rosa26LSL-DTA/+ and Foxc2iCreERT2/+;Rosa26LSL-DTA/+ mice (n=3). Values, mean ± s.e.m.; p-values were obtained using two-tailed t-tests (***p-value <0.001, ****p-value <0.0001).
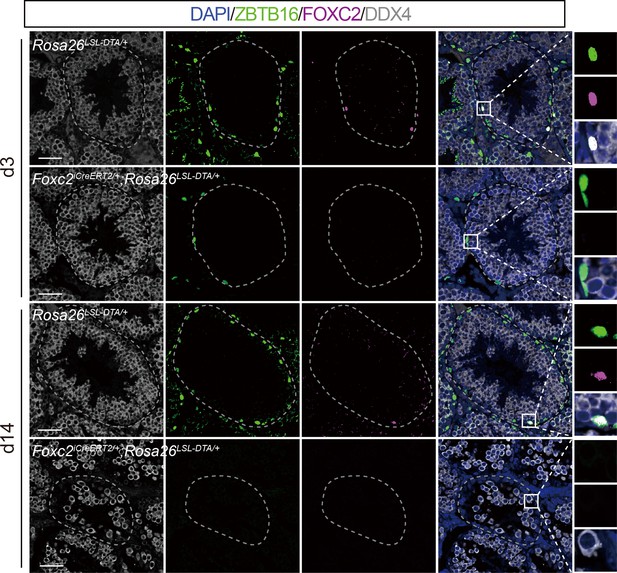
Depletion of undifferentiated spermatogonia (uSPG) pool in Foxc2iCreERT2/+;Rosa26LSL-DTA/+ mice 14 days after specific ablation of FOXC2+ spermatogonial stem cells (SSCs).
Immunostainings for DAPI (blue), DDX4 (white), ZBTB16 (green), and FOXC2 (magenta) at day 3 and day 14 post TAM induction in Foxc2iCreERT2/+;Rosa26LSL-DTA/+ mice (scale bar, 50 μm).
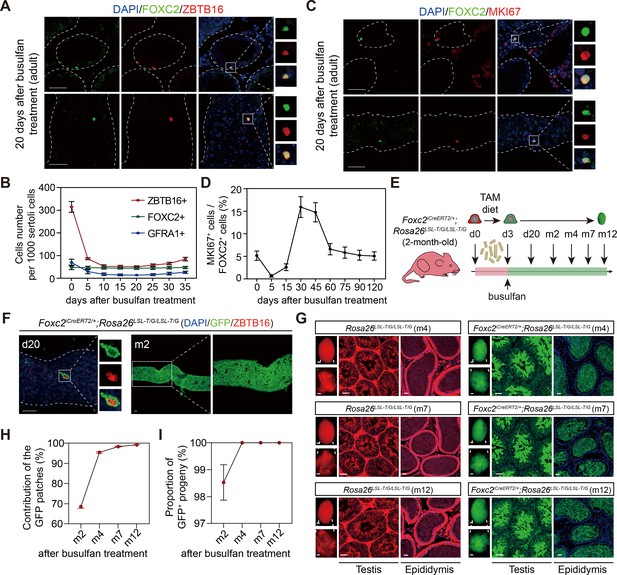
FOXC2+ spermatogonial stem cells (SSCs) are critical for germline regeneration.
(A) Co-immunostaining of FOXC2 (green) with ZBTB16 (red) in seminiferous tubules of the adult testes at day 20 post busulfan treatment. Scale bar, 50 μm. (B) ZBTB16+, GFRA1+, and FOXC2+ population dynamics after busulfan treatment (20 mg/kg, n=10). (C) Co-immunostaining of FOXC2 (green) with MKI67 (red) in seminiferous tubules of the adult testes at day 20 post busulfan treatment. Scale bar, 50 μm. (D) MKI67+FOXC2+ proportions in relation to the whole FOXC2+ population at different time points after busulfan treatment (n=4). (E) Schematic illustration for lineage tracing of FOXC2+ cell after busulfan treatment. (F) Lineage tracing of the GFP+ cells at day 20 and month 2 after busulfan treatment (scale bar, 50 μm). (G) The testes (scale bar, 1 mm), seminiferous tubules, and epididymis (scale bar, 50 μm) at months 4, 7, and 12 post TAM induction and busulfan injection. m, month. (H, I) The proportion dynamics of GFP patches (H) and GFP+ progenies (I). Values, mean ± s.e.m. (n=10). w, week; m, month.
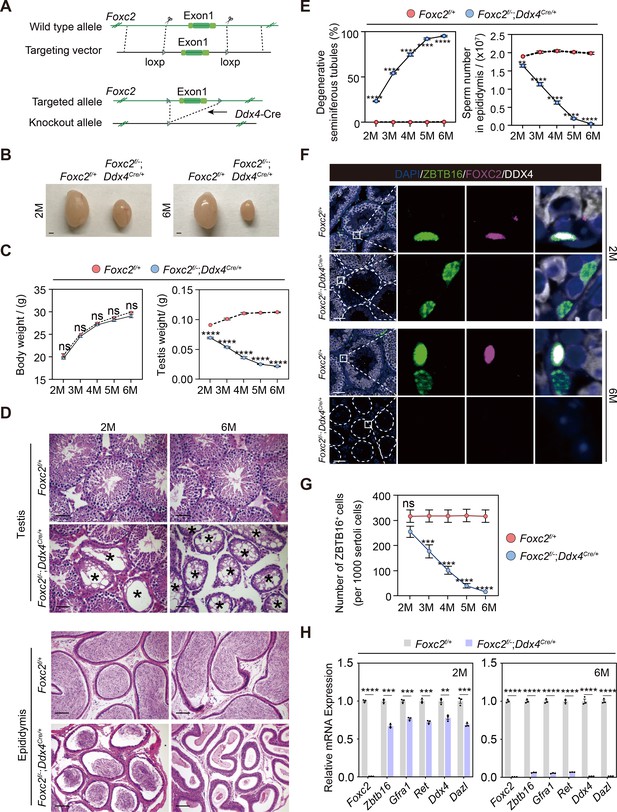
Spermatogenesis exhaustion in the adult Foxc2f/-;Ddx4Cre/+ mice.
(A) Construction of the Foxc2f/-;Ddx4Cre/+ mice. (B) The testes size of the Foxc2f/-;Ddx4Cre/+ mice. Scale bar, 1 mm; M, month. (C) Body weight and testis weight of the Foxc2f/-;Ddx4Cre/+ mice at different age (n=5). M, month; values, mean ± s.e.m.; p-values were obtained using two-tailed t-tests (ns >0.05, ****p-value <0.0001). (D) Hematoxylin and eosin (HE) staining of the testis and epididymis. Scale bar, 50 μm; M, month. (E) Estimation of degenerative tubules and sperm counts in cauda epididymis of the Foxc2f/+ and Foxc2f/-;Ddx4Cre/+ mice with age (n=5). Values, mean ± s.e.m.; p-values were obtained using two-tailed t-tests (**p-value <0.01, ****p-value <0.0001). (F) Immunostainings for DAPI (blue), ZBTB16 (green), FOXC2 (magenta), and DDX4 (white) in the seminiferous tubules of the Foxc2f/+ and Foxc2f/-;Ddx4Cre/+ mice. Scale bar, 50 μm. (G) Estimation of ZBTB16+ uSPG number in the Foxc2f/+ and Foxc2f/-;Ddx4Cre/+ mice with age (n=5). Values, mean ± s.e.m.; p-values were obtained using two-tailed t-tests (ns >0.05, ***p-value <0.001, ****p-value <0.0001). (H) Quantitative RT-PCR analysis of the uSPG and germ cell markers expressed in the testis of the Foxc2f/+ and Foxc2f/-;Ddx4Cre/+ mice (n=3). M, month; values, mean ± s.e.m.; p-values were obtained using two-tailed t-tests (**p-value <0.01, ***p-value <0.001, ****p-value <0.0001).
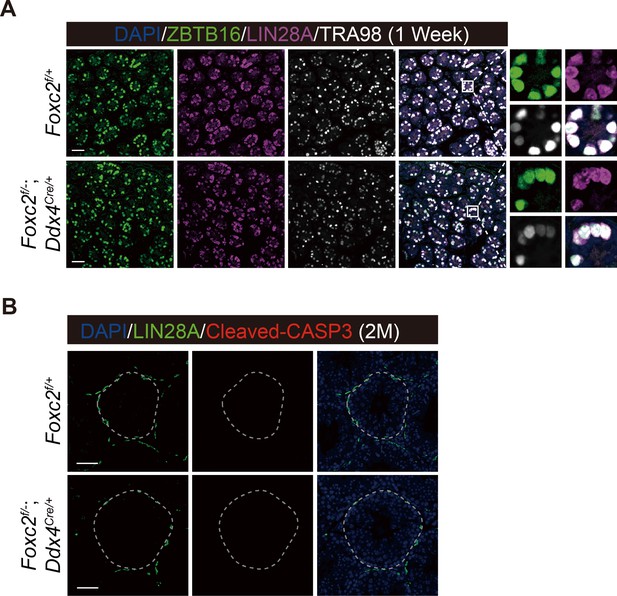
Phenotypic validation of the Foxc2f/-;Ddx4Cre/+ mice.
(A) Immunostainings for DAPI (blue), ZBTB16 (green), LIN28A (magenta), and TRA98 (white) in seminiferous tubules of 1-week-old Foxc2f/+ and Foxc2f/-;Ddx4Cre/+ mice. Scale bar, 50 μm. (B) Immunostainings for DAPI (blue), LIN28A (green), and Cleaved-CASP3 (red) in seminiferous of the Foxc2f/+ and Foxc2f/-;Ddx4Cre/+ mice (2-month-old). M, month; scale bar, 50 μm.
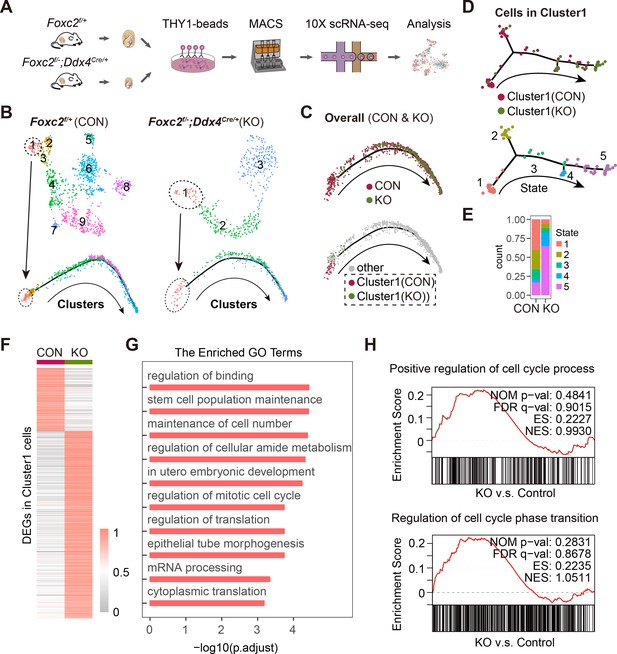
Single-cell RNA-sequencing (scRNA-seq) analysis of THY1+ undifferentiated spermatogonia (uSPG) in Foxc2f/+ and Foxc2f/-;Ddx4Cre/+ mice.
(A) Schematic illustration of the scRNA-seq workflow. (B) t-SNE plot and developmental trajectory of uSPG from Foxc2f/+ and Foxc2f/-;Ddx4Cre/+ mice, respectively, colored by cluster. (C) Developmental trajectories of uSPG from Foxc2f/+ and Foxc2f/-;Ddx4Cre/+ mice, colored by sample or derivation. (D) Developmental trajectories of the cells in Cluster1 from Foxc2f/+ (CON) and Foxc2f/-;Ddx4Cre/+ (KO) mice, colored by derivation or developmental state. (E) The Cluster1 cells proportion of each state in CON and KO mice. (F) Heatmap showing the differentially expressed genes (DEGs) in the Cluster1 cells from the Foxc2f/-;Ddx4Cre/+ mice compared with the Foxc2f/+ mice. (G) Top Gene Ontology (GO) terms enrichment by the down-regulated DEGs in KO mice. (H) Gene set enrichment analysis (GSEA) of the Cluster1 cells (Foxc2f/-;Ddx4Cre/+ v.s. Foxc2f/+ mice). NOM, nominal; FDR, false discovery rate; ES, enrichment score; NES, normalized enrichment score.
-
Figure 6—source data 1
Excel spreadsheet with the list of the differentially expressed genes found by single-cell RNA-sequencing (scRNA-seq) and enriched Gene Ontology terms.
- https://cdn.elifesciences.org/articles/85380/elife-85380-fig6-data1-v1.xlsx
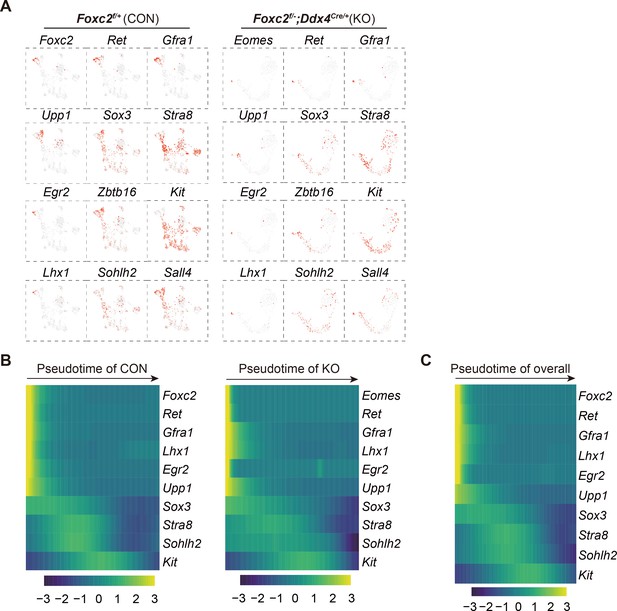
Single-cell RNA-sequencing (scRNA-seq) analysis of THY1+ undifferentiated spermatogonia (uSPG) in adult Foxc2f/+ and Foxc2f/-;Ddx4Cre/+ mice.
(A) Feature plots of classic SPG markers for uSPG in adult Foxc2f/+ or Foxc2f/-;Ddx4Cre/+ mice. (B) Expression dynamics of SPG markers with pseudotime progression for uSPG from Foxc2f/+ or Foxc2f/-;Ddx4Cre/+ mice, respectively. (C) Expression dynamics of SPG markers with pseudotime progression for overall uSPG from Foxc2f/+ and Foxc2f/-;Ddx4Cre/+ mice.
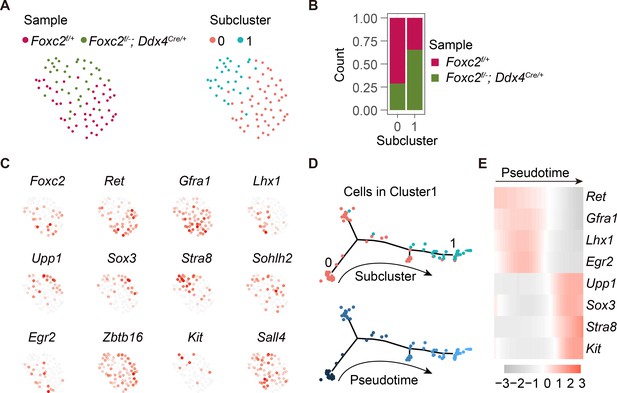
Re-cluster and developmental trajectory analysis of cells in Cluster1 derived from adult Foxc2f/+ and Foxc2f/-;Ddx4Cre/+ mice.
(A) The t-SNE plot of the Cluster1 cells aggregated from the Foxc2f/+ and Foxc2f/-;Ddx4Cre/+ mice colored by sample or subcluster. (B) The cell proportion of each sample in each subcluster. (C) Feature plots of SPG markers expression. (D) Developmental trajectory of the aggregated Cluster1 cells colored by subcluster or pseudotime. (E) Expression dynamics of SPG markers with pseudotime progression.
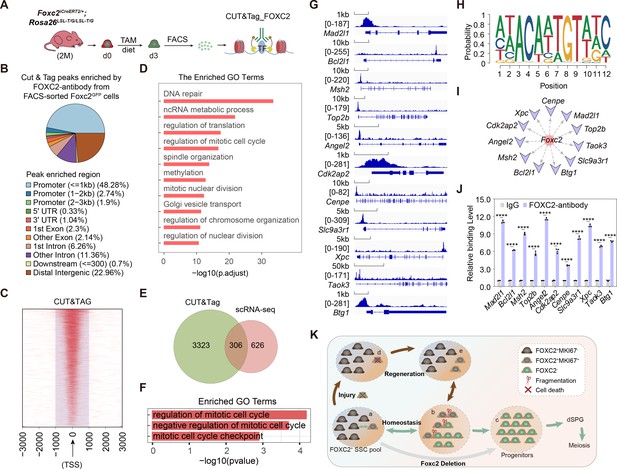
FOXC2 is essential for sustaining the quiescent state of spermatogonial stem cells (SSCs) via regulating cell cycle.
(A) Workflow schematic illustration of the CUT&Tag_FOXC2 analysis on the fluorescence-activated cell sorting (FACS)-sorted FOXC2+ cells. (B) Pie chart for CUT&Tag_FOXC2 peaks genome distribution. (C) Profiling of CUT&Tag_FOXC2 peaks in proximity to transcriptional starting site (TSS). The distance to TSS within 1000 was highlighted in the purple box. (D) Top Gene Ontology (GO) terms enrichment by genes annotated by CUT&Tag_FOXC2 peaks. (E) Venn diagram of FOXC2 target genes defined by overlapping the CUT&Tag sequencing and single-cell RNA-sequencing (scRNA-seq) datasets. (F) GO terms enrichment by the FOXC2 target genes related to cell cycle regulation. (G) Chromatin landscapes of CUT&Tag_FOXC2 peaks of the candidates associated with negative cell cycle regulation. (H) The DNA-binding motif for FOXC2 (predicted with HOMER). (I) The cell cycle-related candidates possessing high binding potential (>0.8, predicted with JASPAR SCAN). (J) CUT&Tag-qPCR validation of the cell cycle arrest regulatory genes. (n=3). Values, mean ± s.e.m.; p-values were obtained using two-tailed t-tests (****p-value <0.0001). (K) The model for the maintenance of the FOXC2+ SSC subpopulation in adult testis.
-
Figure 7—source data 1
Excel spreadsheet with the list of the differentially expressed genes found by Cleavage Under Targets and Tagmentation (CUT&Tag) sequencing and Gene Ontology terms of the 306 crossed candidates.
- https://cdn.elifesciences.org/articles/85380/elife-85380-fig7-data1-v1.xlsx
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85380/elife-85380-mdarchecklist1-v1.docx
-
Supplementary file 1
Excel spreadsheet with the primers and antibodies used in this study.
- https://cdn.elifesciences.org/articles/85380/elife-85380-supp1-v1.xlsx