A parabrachial to hypothalamic pathway mediates defensive behavior
Figures
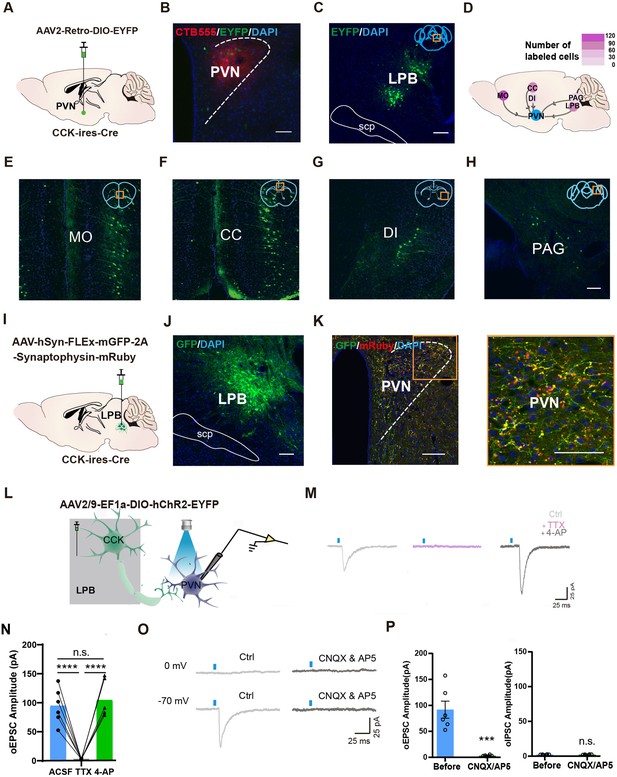
LPBCCK neurons project to the PVN.
(A) Scheme of viral strategy for retrograde tracing from the PVN in Cck-cre mice using AAV2-Retro virus. (B) Representative image showing the injection site as marked by CTB555. (C) Representative histological images of EYFP+ neurons in the LPB. (D–H) A heatmap (D) demonstrating the distribution of EYFP+ neurons in the MO (E), CC (F), DI (G), PAG (H), n = 3 mice. (I–J) Anterograde tracing of LPBCCK neurons using AAV-hSyn-FLEX-mGFP-2A-Synaptophysin-mRuby virus. (K) Projections of LPBCCK neurons to the PVN. The right panel shows a magnified view of boxed area; scale bar, 100 μm; n = 3 mice. (L) Schematic of recording from PVN cells after optogenetic activation of LPBCCK axonal terminals. (M) Representative traces of light-evoked EPSCs recorded from PVN neurons following light stimulation of LPBCCK axonal terminals in the presence of ACSF (Ctrl), TTX (1 μM) and 4-AP (100 μM). (N) Quantification of excitatory postsynaptic currents (EPSCs) from identified PVN neurons receiving inputs from the LPBCCK neurons in the presence of ACSF (Ctrl), TTX (1 μM) and 4-AP (100 μM). (ACSF vs. TTX ****p < 0.0001; TTX vs. 4-AP ****p < 0.0001; ACSF vs. 4-AP p > 0.9999, one-way ANOVA Bonferroni’s multiple comparisons test). (O) Representative traces of light-evoked EPSCs recorded from PVN neurons following light stimulation of LPBCCK axonal terminals in the presence of CNQX (20 μM) and AP5 (50 μM) (n = 9 neurons). (P) Quantification of EPSCs and inhibitory postsynaptic currents (IPSCs) from identified PVN neurons receiving inputs from the LPBCCK neurons. (oEPSC, p = 0.0003 t = 5.378 df = 10; oIPSC, p = 0.8793 t = 0.1558 df = 10; unpaired t test).
-
Figure 1—source data 1
Quantification of the labeled cells, EPSCs and IPSCs.
- https://cdn.elifesciences.org/articles/85450/elife-85450-fig1-data1-v1.xlsx
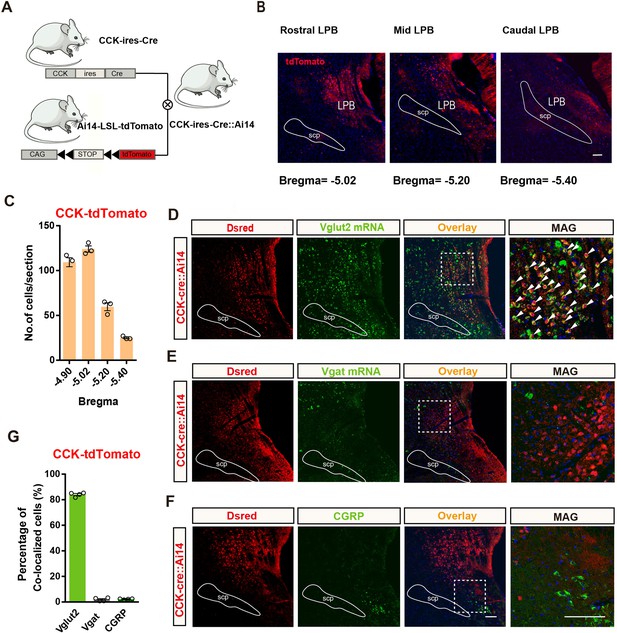
Distribution and identification of CCK neurons in the LPB.
(A) Schematic of mice crossing to generate the Cck-ires-cre::Ai14 mice. (B) Representative coronal section images illustrating the expression of CCK-tdT in the LPB; scale bar, 100 μm. (C) Quantification of CCK-tdT cells across the rostral-to-caudal extent of the LPB; n = 3 mice. (D) Representative images showing co-labeling (arrowheads) of Vglut2-positive (Green) and CCK-tdT expressing cells in the LPB. Rightmost panel, magnification of the box-shaped area in the left panel (white dashed lines). Blue, DAPI staining. Scale bar, 100 μm. (E) Example images showing Vgat-positive (Green) and CCK-tdT-expressing cells in the LPB. Rightmost panel, magnification of the boxed area in the left panel. Blue, DAPI staining. Scale bar, 100 μm. (F) Representative image from a Cck-ires-cre::Ai14 mouse in the LPB region stained with antibodies against CGRP. Rightmost panels, magnification of the boxed areas in the left panels. Blue, DAPI staining. Scale bar, 100 μm. (G) Percentage of CCK+ neurons that co-localized with markers; n = 4 mice.
-
Figure 1—figure supplement 1—source data 1
Quantification of LPB CCK-tdT cells.
- https://cdn.elifesciences.org/articles/85450/elife-85450-fig1-figsupp1-data1-v1.xlsx
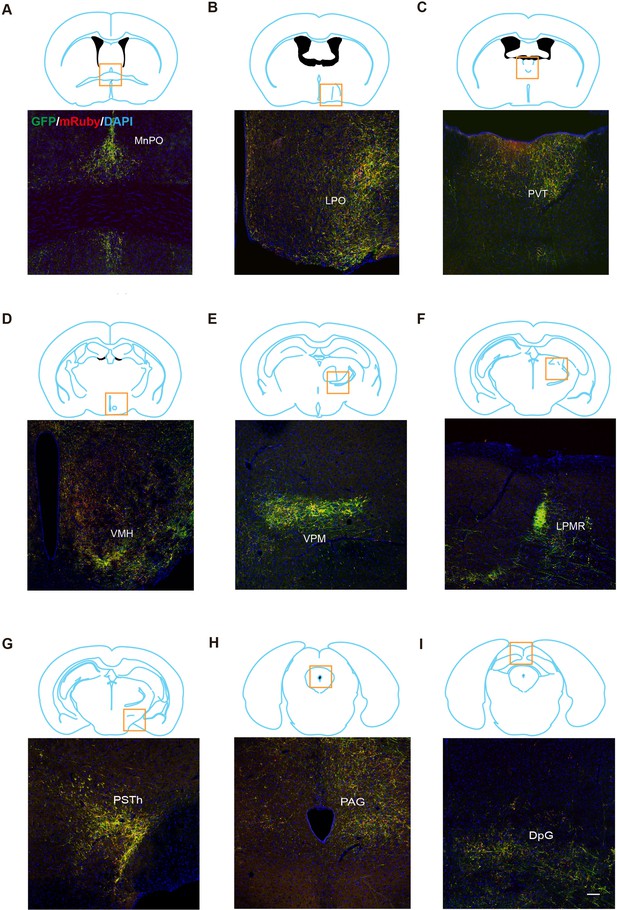
Anterograde mapping of LPBCCK neurons.
(A–I) Representative images showing the projections from LPBCCK neurons in the median preoptic nucleus (MnPO), lateral preoptic area (LPO), paraventricular thalamic nucleus (PVT), ventromedial hypothalamic nucleus (VMH), ventral posteromedial thalamic nucleus (VPM), lateral posterior thalamic nucleus, mediorostral part (LPMR), parasubthalamic nucleus (PSTh), periaqueductal gray (PAG), deep gray layer of the superior colliculus (DpG)(n=3 mice; green, GFP; red, mrubby; blue, DAPI). Scale bar, 100 μm; n = 3 mice.
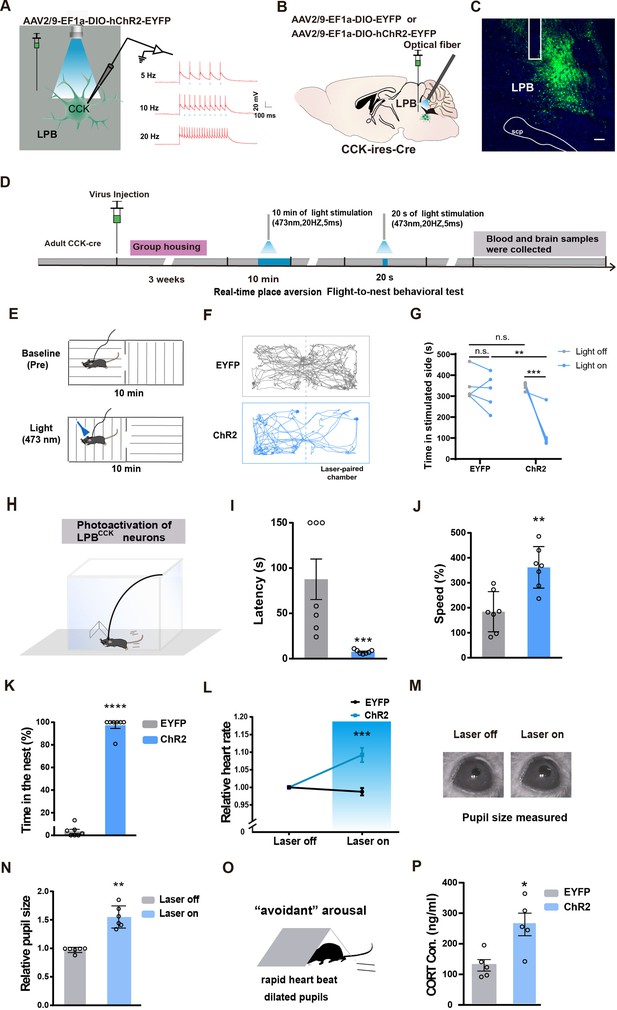
Activation of LPBCCK neurons triggers aversion, defensive-like flight-to-nest behavior and autonomic responses.
(A) Left, schematic of light stimulation of and patch-clamp recording from ChR2-EYFP expressing CCK neurons in the LPB. Right, example of action potentials evoked by optogenetic stimulation LPBCCK neurons using whole cell patch-clamp slice recording. (B) Schematic diagram of optogenetic activation of LPBCCK neurons. (C) Representative image showing the ChR2-EYFP expression and optical fiber tip locations in the LPB of a Cck-cre mouse. Scale bar, 100 μm. (D) Schematic of the timing and behavioral paradigm with optical activation of LPBCCK neurons. (E–F) Diagram of the real-time place aversion (RTPA) test and the example traces of the RTPA test from the mice. (G) Quantification of the time of mice spent in the laser-paired chamber (EYFP: n = 5 mice, ChR2: n = 5 mice; df = 16; two-way ANOVA test) after optogenetic activation. (H) Diagram of the flight to nest test. (I–K) Quantification of latency (I), speed (J) and time in the nest (K) (EYFP: n = 7 mice, ChR2: n = 7 mice; for latency, p = 0.0006, U = 0; Mann-Whitney test; for speed, p = 0.0016, t = 4.052, df = 12; unpaired t test; for time in the nest, p < 0.0001, t = 19.82, df = 12; unpaired t test). (L) Analyses of heart rate changes induced by photostimulation of LPBCCK neurons. (EYFP: n = 7 mice, ChR2: n = 7 mice; p = 0.0006, t = 4.603, df = 12; unpaired t test). (M–N) Example images of computer-detected pupils (M) and quantitative analyses of pupil size before and during photo-stimulation of LPBCCK neurons (N) (EYFP: n = 6 mice, ChR2: n = 6 mice; p = 0.0022, U = 0; Mann-Whitney test). (O) Cartoon of the arousal state during the activation of LPBCCK neurons. (P) Plasma corticosterone levels in EYFP and ChR2 groups. (EYFP: n = 5 mice, ChR2: n = 5 mice; p = 0.0121, t = 3.23, df = 8; unpaired t test).
-
Figure 2—source data 1
Quantification of the flight-to-nest behavior and autonomic responses upon activation of LPB CCK neurons.
- https://cdn.elifesciences.org/articles/85450/elife-85450-fig2-data1-v1.xlsx
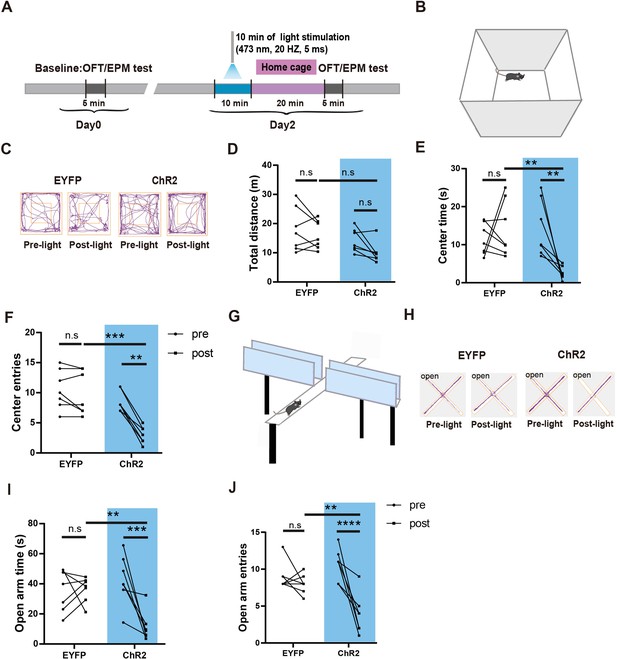
Photostimulation of LPBCCK neurons induces anxiety-like behaviors.
(A) Schematic of behavioral test protocol. OFT, open-field test; EPM, elevated plus maze test. (B) Experimental apparatus: Open field. (C) Example movement traces of a control (AAV2/9-EF1a-DIO-EYFP, left) and an experimental mouse (AAV2/9-EF1a-DIO-ChR2-EYFP, right) in the OFT. (D–F) Quantification of the total distance moved, time spent in the center and total number of center entries in the OFT before (Pre) and after (Post) photostimulation of LPBCCK neurons in Cck-cre mice (EYFP and ChR2, n = 7 mice; for total distance, Pre- light vs. Post-light EYFP p>0.9999, Post-light EYFP vs. ChR2, p = 0.2236, Pre-light vs. Post-light ChR2, p = 0.9026; two-way ANOVA, Bonferroni’s multiple comparisons test; for center time, Pre-light vs. Post-light EYFP, p > 0.9999, Post-light EYFP vs. Post-light ChR2 **p < 0.01 p = 0.005, Pre-light ChR2 vs. Post-light ChR2 **p < 0.01 p = 0.005; two-way ANOVA, Bonferroni’s multiple comparisons test; for center entries, Pre-light EYFP vs. Post-light EYFP ns p > 0.9999, Post-light EYFP vs. Post-light ChR2 ***p < 0.0005 p = 0.0004, Pre-light ChR2 vs. Post-light ChR2 **p < 0.01 p = 0.0042; two-way ANOVA, Bonferroni’s multiple comparisons test). (G) Experimental apparatus: Elevated plus maze. (H) Example traces of a control group mouse (AAV2/9-EF1a-DIO-EYFP, left) and an experimental group mouse (AAV2/9-EF1a-DIO-EYFP, right) in the elevated plus maze. (I–J) Quantification of the open arm time and entries in the elevated plus maze tests before (Pre) and after (Post) photostimulation of LPBCCK neurons in Cck-cre mice. (EYFP: n = 7 mice, ChR2: n = 7 mice; for open arm time, Pre-light EYFP vs. Post-light EYFP ns p > 0.9999, Post-light EYFP vs. Post-light ChR2 **p < 0.01 p = 0.0053, Pre-light ChR2 vs. Post-light ChR2 ***p < 0.0005 p = 0.0005; two-way ANOVA, Bonferroni’s multiple comparisons test; for open arm entries, Pre-light EYFP vs. Post-light EYFP ns p > 0.9999, Post-light EYFP vs. Post-light ChR2 **p < 0.01 p = 0.0159, Pre-light ChR2 vs. Post-light ChR2 ****p < 0.0001; two-way ANOVA, Bonferroni’s multiple comparisons test).
-
Figure 2—figure supplement 1—source data 1
Quantification of the anxiety-like behavior.
- https://cdn.elifesciences.org/articles/85450/elife-85450-fig2-figsupp1-data1-v1.xlsx
Images of ChR2-EYFP expression in the LPB and optical fiber implantation above the LPB (A-G), with circles (H-I) indicating the location of optical fibers.
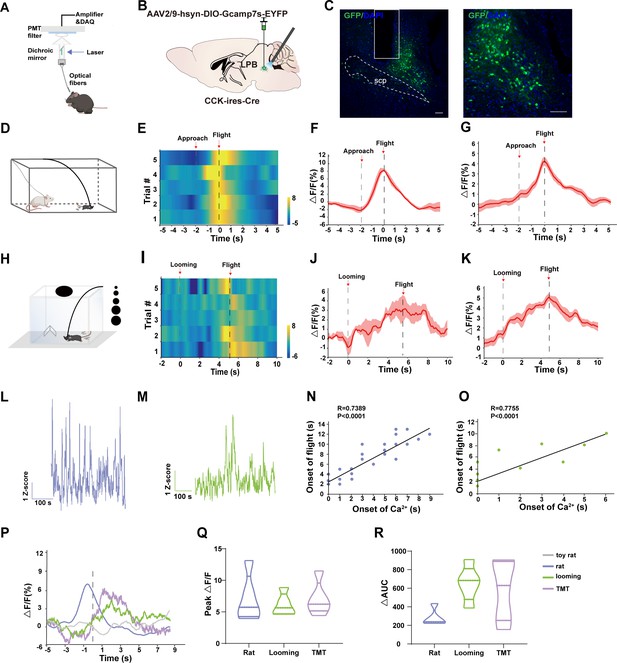
Threat stimuli recruit LPBCCK neurons to elicit defensive behaviors.
(A) Schematic of the in vivo recording system for the Ca2+ signal. (B) Schematic showing the injections and recording of LPB neurons in Cck-cre mice. (C) A representative image (left panel) and a magnified view of neurons labeled by AAV-hsyn-DIO-GCaMP7s, scale bar, 100 μm; n = 5 mice. (D) Schematic of the rat exposure assay. (E–F) A heatmap (E) and a peri-event plot (F) of calcium transients of LPBCCK neurons in a mouse evoked by rat exposure (5 trials) (gray dotted line, onset of approach to the live rat; dark dotted line, onset of flight). (G) Average calcium transients of the tested animals during the rat exposure assay (n = 5 mice). Shaded areas around means indicate error bars. (H) Schematic paradigm of looming stimulus in a nest-containing open-field apparatus. (I–J) A heatmap presentation (I) and a peri-event plot (J) of calcium transients of LPBCCK neurons in a mouse upon looming stimulus (5 trials). (K) Average calcium transients of the tested animals during the looming assay (n = 5 mice). (L–M) Long session calcium recordings of LPBCCK neurons during rat exposure (L) or looming tests (M). (N–O) Correlation analyses between the elevation of calcium transients and the onset of flight during rat exposure or looming tests. (p < 0.0001 Linear regression). (P) Comparison of calcium signals LPBCCK neurons evoked by different stimuli. (Q–R) Plot depicting the differences of the amplitude or the area under the curve (AUC) of calcium signal changes in response to different stimuli. ∆AUC, AUC stimulus signal- AUC basal signal. (Q, Rat vs. Looming: p > 0.9999; Rat vs. TMT: p > 0.9999;TMT vs. Looming: p > 0.9999; one-way ANOVA; R, Rat vs. Looming p > 0.0676; Rat vs. TMT p > 0.1654; TMT vs. Looming p >0.9999; one-way ANOVA).
-
Figure 3—source data 1
Plot depicting the difference of Ca2+ activity.
- https://cdn.elifesciences.org/articles/85450/elife-85450-fig3-data1-v1.xlsx
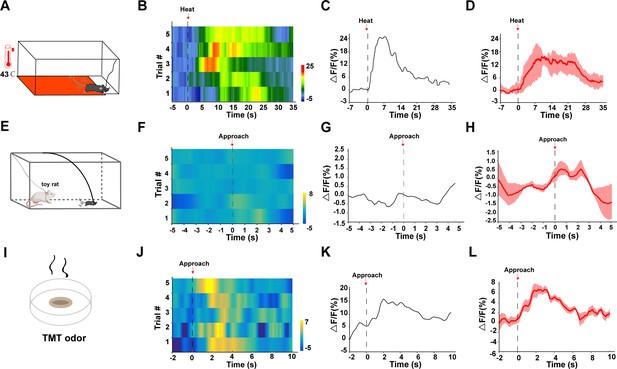
LPBCCK neurons respond to threat stimuli.
(A) A schematic of the heat assay. (B) Representative trial-by-trial heatmap presentation of calcium transients evoked by heat exposure in five trials from one mouse. (C) Representative peri-event plot of the one trial of calcium transients. (D) Average calcium transients for the five trials from one mouse. (E) Toy rat assay schematic. Mice were placed in the presence of a toy rat. (F) Representative trial-by-trial heatmap presentation of calcium transients evoked by toy rat exposure in one trial from one mouse. (G) Representative peri-event plot of the five trials of calcium transients. (H) Average calcium transients for the entire test group. (I) TMT odor exposure schematic. (J) Representative trial-by-trial heatmap presentation of calcium transients evoked by rat exposure in one trial from one mouse. (K) Representative peri-event plot of the five trials of calcium transients. (L) Average calcium transients for the entire test group.
Images of AAV-DIO-synapse-jGCaMP7b virus infection at the LPB (A) and downstream PVN and PSTh (B-C).
Images of Gcamp7s expression in the LPB and optical fiber implantation in the LPB (A-E), with circles (F-G) indicating the location of optical fibers.
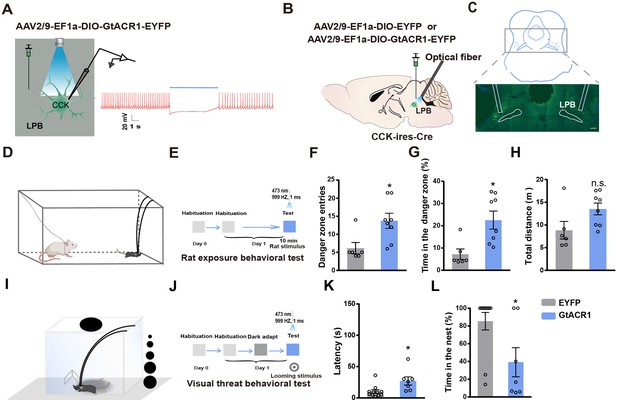
Optogenetic inhibition of LPBCCK neurons suppresses predator- and visual predatory cue-evoked innate flight responses.
(A) Left, schematic of light stimulation of and patch-clamp recording of GtACR1-expressing CCK neurons in the LPB. Right, example of action potentials evoked by optogenetic inhibition of LPBCCK neurons from using whole cell patch-clamp recording. (B) Schematic diagram of optogenetic inhibition of LPBCCK neurons. (C) Representative image showing the GtACR1-EYFP expression in the LPB and optical fiber tip locations above the LPB of a Cck-cre mouse. (D–E) Schematic and the timing and behavioral paradigm of rat exposure assay. (F–H) Photoinhibition of LPBCCK neurons increased number of entries toward the rat (danger zone), time spent in the danger zone, with unchanged travel distance (EYFP: n = 6 mice, GtACR1: n = 9 mice; for times of entries, p = 0.0456, t = 2.21, df = 13; unpaired t test; for time in the danger zone, p = 0.0153, t = 2.791, df = 13; unpaired t test; for total distance, p = 0.1246, t = 1.642, df = 13; unpaired t test). (I–J) Schematic of the looming test apparatus, the timing and behavioral paradigm of looming-evoked flight-to-nest behavioral test. (K–L) Photoinhibition of LPBCCK neurons increased the latency towards the nest and reduced the hiding time in the nest. (EYFP: n = 11 mice, GtACR1: n = 7 mice; for latency, p = 0.0153, t = 2.716, df = 16; unpaired t test; Mann-Whitney test; for time in the nest, p = 0.0201, t = 2.582, df = 16; unpaired t test).
-
Figure 4—source data 1
Quantification of the defensive responses upon inhibition of LPB CCK neurons.
- https://cdn.elifesciences.org/articles/85450/elife-85450-fig4-data1-v1.xlsx
Images of GtACR1-EYFP expression in the LPB and optical fiber implantation above the LPB (A-I), with circles (J-L) indicating the location of optical fibers.
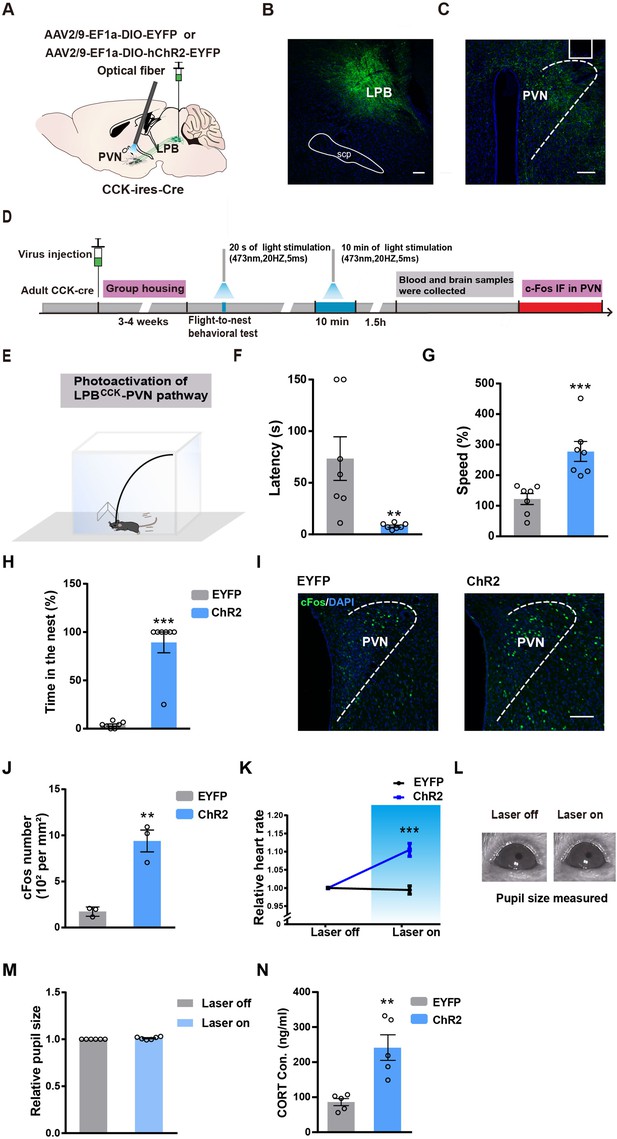
Photostimulation of the LPBCCK-PVN pathway induces defensive-like flight-to-nest behavior.
(A) Schematic diagram of optogenetic activation of LPBCCK-PVN pathway. (B–C) A representative image showing the ChR2-EYFP expression in the LPB (B) and the optical fiber tip locations in the PVN (C) of a Cck-cre mouse. (D) Schematic of the timing and behavioral paradigm with optical activation of LPBCCK-PVN pathway. (E) Schematic of the experimental apparatus with a nest in the corner. (F–H) Optogenetic activation of the LPBCCK-PVN pathway shortened the latency but increased the speed of animals towards the nest, with increased hiding time in the nest (EYFP: n = 7 mice, ChR2: n = 7 mice; for latency, p = 0.0012, U = 0; Mann-Whitney test; for speed, p = 0.0013, t = 4.163, df = 12; unpaired t test; for time in the nest, p = 0.0006, U = 0; Mann-Whitney test). (I–J) C-fos staining in the PVN (I) and quantification of c-fos positive cells in the PVN (J). Scale bar: 100 μm. (EYFP: n = 3 mice, ChR2: n = 3 mice; p = 0.0033, t = 6.253, df = 4; unpaired t test). (K) Mean heart rate analyses in EYFP and ChR2 groups (EYFP: n = 7 mice, ChR2: n = 7 mice; p = 0.0002, t = 5.249, df = 12; unpaired t test). (L) Example image of computer-detected pupil size before and during photoactivation of LPBCCK neurons. (M) Relative pupil size of animals (during/before photostimulation of LPBCCK neurons) (EYFP: n = 6 mice, ChR2: n = 6 mice; p = 0.0022, U = 0; Mann-Whitney test). (N) Plasma corticosterone levels in EYFP and ChR2 groups (EYFP: n = 5 mice, ChR2: n = 5 mice; p = 0.0079, U = 0; Mann-Whitney test).
-
Figure 5—source data 1
Quantification of the flight-to-nest behavior and autonomic responses upon activation of LPB CCK-PVN pathway.
- https://cdn.elifesciences.org/articles/85450/elife-85450-fig5-data1-v1.xlsx
Images of ChR2-EYFP expression in the LPB and optical fiber implantation above the PVN (A-F).
The square boxes indicate the location of virus expression and circles represent optical fiber location (G-L).
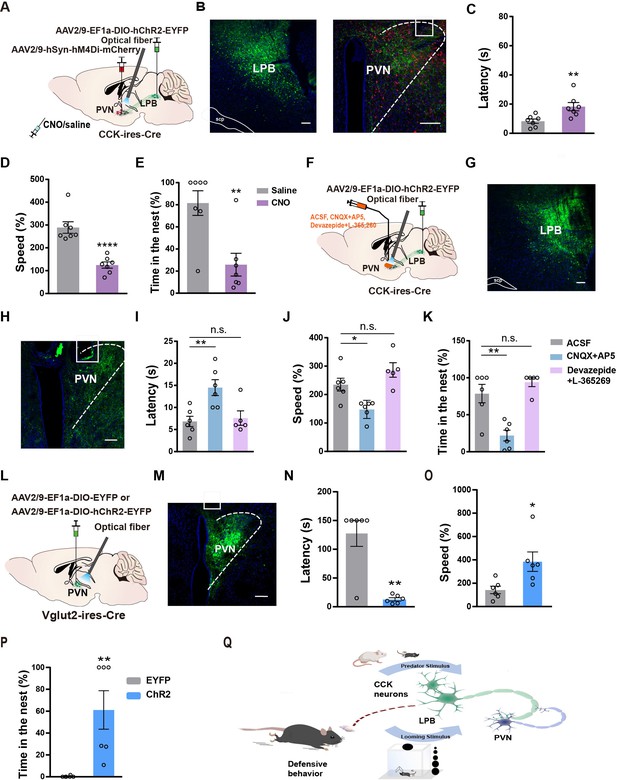
PVN is involved in the defensive-like flight-to-nest behavior evoked by LPBCCK neurons.
(A) Schematic of experimental setup. (B) Left, representative image showing the ChR2-EYFP expression in the LPB of a Cck-cre mouse. Scale bar, 100 μm. Right, representative image showing the hM4Di-mCherry expression in the PVN and optic fiber placement in the PVN from a ChR2-EYFP expressing mouse. (C–E) Chemogenetic inhibition of PVN neurons before optical activation of LPBCCK-PVN terminals increased latency and reduced the speed of animals towards the nest, with reduced hiding time in the nest (saline: n = 7 mice, CNO: n = 7 mice; for latency, p = 0.0088, t = 3.122, df = 12; unpaired t test; for speed, p < 0.0001, t = 5.736, df = 12; unpaired t test; for time in the nest, p = 0.0032, t = 3.665, df = 12; unpaired t test). (F) Optogenetic activation of LPBCCK -PVN terminals with a cannula implanted in the PVN for aCSF, CNQX +AP5, or Devazepide +L-365 260 delivery. Scale bar, 100 μm. (G) Representative image showing the ChR2-EYFP expression in the LPB of a Cck-cre mouse. Scale bar, 100 μm. (H) Representative image showing the cannula placement in the PVN from a ChR2-EYFP expressing Cck-cre mouse. Scale bar, 100 μm. (I–K) Microinjection of the glutamate receptor antagonists (CNQX +AP5), rather than CCK receptor antagonists (Devazepide +L-365260), into the PVN increased the latency to the nest and reduced the hiding time in the nest (ACSF: n = 6 mice, CNQX +AP5: n = 6 mice, Devazepide +L-365260: n = 5 mice; for latency, F(2,14) = 7.658, p = 0.057; One-way ANOVA; for speed, F(2,14) = 11.49, p = 0.0011; for time in the nest, One-way ANOVA; F(2,14) = 16.44, p = 0.0002; One-way ANOVA). (L) Schematic diagram of optogenetic activation of PVNVglut2 neurons. (M) Representative image showing the ChR2-EYFP expression and optical fiber tip locations in the PVN of a Vglut2-cre mouse. Scale bar, 100 μm. (N–P) Optogenetic activation of PVNVglut2 neurons reduced the latency, increased the speed of animals towards a nest and the time in the nest. (EYFP: n = 6 mice, ChR2: n = 6 mice; for latency, p = 0.0065, U = 2; Mann-Whitney test; for speed, p = 0.0221, t = 2.705, df = 10; unpaired t test; for time in the nest, p = 0.022, U = 0; Mann-Whitney test). (Q) Graphical summary showing the LPBCCK-PVN pathway in mediating defensive behaviors.
-
Figure 6—source data 1
Quantification of the flight-to-nest behavior upon manipulation of PVN neurons.
- https://cdn.elifesciences.org/articles/85450/elife-85450-fig6-data1-v1.xlsx
-
Figure 6—source data 2
Maps for virus expression and optical fiber location.
- https://cdn.elifesciences.org/articles/85450/elife-85450-fig6-data2-v1.docx
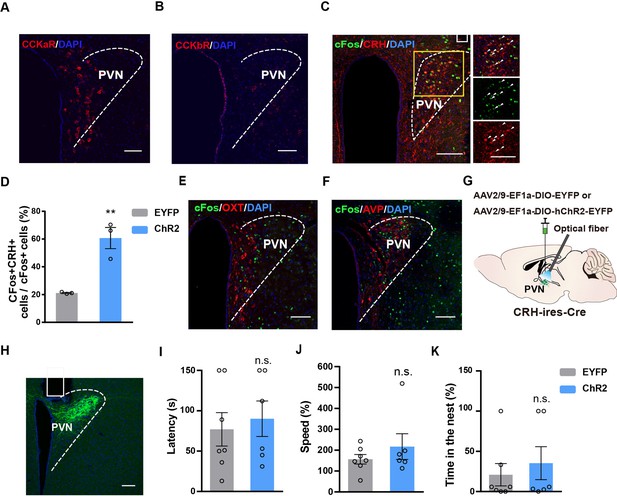
Stimulation of LPBCCK-PVN pathway activates PVNCRH neurons, but activation of PVNCRH neurons does not trigger flight-to-nest behavior.
(A–B) Representative histological images showing Cckar and Cckbr in the PVN. (C) C-fos expression and colocalization (arrows) with CRH in the PVN with blue light activation of LPB-originating ChR2-EYFP expressing terminals in the PVN. Scale bar: 100 μm. (D) Quantification for the percentage of activated CRH neurons in the PVN following blue light activation. (EYFP: n = 3 mice, ChR2: n = 3 mice; p = 0.0066, t = 5.187, df = 4; unpaired t test). (E–F) Left: C-fos expression and colocalization (arrows) with OXT in the PVN with blue light activation of LPB-originating ChR2-EYFP expressing terminals in the PVN. Scale bar: 100 μm. Right: C-fos expression and colocalization (arrows) with AVP in the PVN with blue light activation of LPB-originating ChR2-EYFP expressing terminals in the PVN. Scale bar: 100 μm. (G) Schematic diagram of optogenetic activation of PVNCRH neurons. (H) Representative image showing the ChR2-EYFP expression and optical fiber tip locations in the PVN of a Crh-cre mouse. Scale bar, 100 μm. (I–K) Optogenetic activation of PVNCRH neurons did not induce flight-to-nest behavior. Compared with control group, ChR2 group showed no changes in latency, speed back into the nest and percentage of hiding time spent in the nest (EYFP: n = 5 mice, ChR2: n = 6 mice; for latency, p = 0.6664, t = 0.4456, df = 9; unpaired t test; for speed, p = 0.0221, t = 2.705, df = 10; unpaired t test; for time in the nest, p = 0.2088, t = 1.367, df = 8; unpaired t test).
-
Figure 6—figure supplement 1—source data 1
Quantification of the CRH neurons and flight-to-nest behavior.
- https://cdn.elifesciences.org/articles/85450/elife-85450-fig6-figsupp1-data1-v1.xlsx
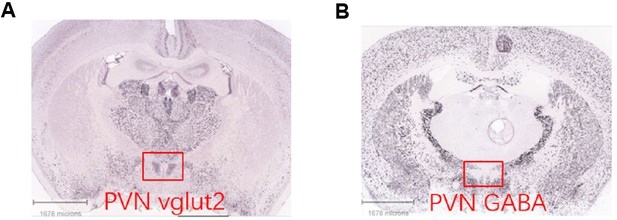
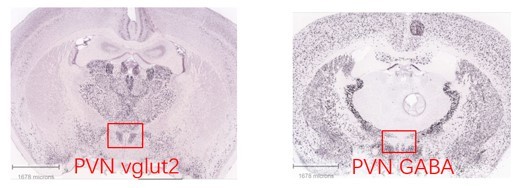
Images of ISH data from the Allen Brain Atlas.
ISH data from the Allen Brain Atlas showed that Vglut2 neurons are located in the PVN (A), whereas GABAergic neurons are almost excluded from the PVN region by surrounding the PVN (B).
Images of ChR2-EYFP expression in the PVN and optical fiber implantation above the PVN (A-F).
The circles represent optical fiber location (G-I).
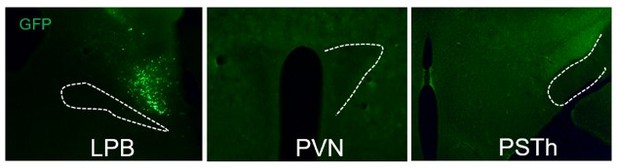
Representative figures of AAV-DIO-synapse-jGCaMP7b infection at the LPB and downstream PVN and PSTh.
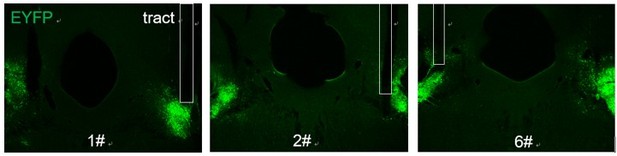
Representative images of fibers implantation above the LPB after injection of AAV-DIO-GtACR1-EYFP virus.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | Cck-ires-cre(Ccktm1.1(cre)Zjh/J) | The Jackson Laboratory | JAX012706 | |
| Strain, strain background (Mus musculus) | Crh-ires-cre (B6(Cg))-Crhtm1(cre)Zjh/J | The Jackson Laboratory | JAX012704 | |
| Strain, strain background (Mus musculus) | Vglut2-ires-Cre (Slc17a6tm2(cre)Lowl/J) | The Jackson Laboratory | JAX 016963 | |
| Strain, strain background (Mus musculus) | Ai14 (B6;129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J) | The Jackson Laboratory | JAX007908 | |
| Strain, strain background (Rattus norvegicus) | SD | Shanghai SLAC Laboratory Animal Co. Ltd | http://www.slaccas.com/ | |
| Strain, strain background (Mus musculus) | C57BL/6 J | Shanghai SLAC Laboratory Animal Co. Ltd | http://www.slaccas.com/ | |
| Antibody | Anti-Dsred, rabbit polyclonal | Takara | Cat# 632496 RRID: AB_10013483 | (1:800) |
| Antibody | Anti-CGRP, mouse monoclonal | Abcam | Cat# 81887 RRID: AB_1658411 | (1:800) |
| Antibody | Anti-GFP, goat polyclonal | Abcam | Cat# 5450 RRID: AB_304896 | (1:500) |
| Antibody | Anti-CRH, rabbit polyclonal | Phoenix Biotech | Cat# H-019–06 | (1:500) |
| Antibody | Anti-c-fos, Guinea pig polyclonal | Synaptic Systems | Cat# 226004 RRID: AB_2619946 | (1:10,000) |
| Antibody | Alexa Fluor 488 donkey anti-guinea pig IgG (H+L) polyclonal | Jackson | Cat#112-486-068 RRID: AB_2617153 | (1:1000) |
| Antibody | Alexa Fluor 555 donkey anti-rabbit IgG (H+L) polyclonal | Invitrogen | Cat#A31572 RRID: AB_162543 | (1:1000) |
| Antibody | Alexa Fluor 488 donkey anti-mouse IgG (H+L) polyclonal | Invitrogen | Cat# R37114 RRID: AB_2556542 | (1:1000) |
| Antibody | Alexa Fluor 488 donkey anti-goat IgG (H+L) polyclonal | Invitrogen | Cat# A11055 RRID: AB_2534102 | (1:1000) |
| Antibody | Alexa Fluor 488 donkey anti-rabbit IgG (H+L) polyclonal | Jackson | Cat# R37118 RRID: AB_2556546 | (1:1000) |
| Commercial assay or kit | RNAscope Multiplex Fluorescent Reagent Kit v2 | Advanced Cell Diagnostics | Cat# 323100 | |
| Sequence-based reagent | RNAscope probe Slc17a6 | Advanced Cell Diagnostics | accession number NM_080853.3 | probe region 1986–2998 |
| Sequence-based reagent | RNAscope probe Slc32a1 | Advanced Cell Diagnostics | Accession number NM_009508.2 | probe region 894–2037 |
| Sequence-based reagent | RNAscope probe Cckar | Advanced Cell Diagnostics | accession number NM_009827.2 | probe region 328–1434 |
| Sequence-based reagent | RNAscope probe Cckbr | Advanced Cell Diagnostics | accession number NM_007627.4 | probe region 136–1164 |
| Chemical compound, drug | CNQX disodium salt hydrate | Sigma-Aldrich | Cat#1045 | |
| Chemical compound, drug | Devazepide | Sigma-Aldrich | Cat#2304 | |
| Chemical compound, drug | Clozapine N-oxide | Sigma-Aldrich | Cat#C0832 | |
| Chemical compound, drug | L-365,260 | Sigma-Aldrich | Cat#143626 | |
| Chemical compound, drug | DAPI | Sigma-Aldrich | N/A | |
| Chemical compound, drug | Tween-20 | Sigma-Aldrich | N/A | |
| Strain, strain background (AAV2/9) | AAV2/9-hEF1a-DIO-hChR2(H134R)-EYFP-WPRE-pA | Shanghai Taitool Bioscience Co. | Cat# S0199-9 | Viral titers: 2.95x1013 particles/ml |
| Strain, strain background (AAV2/9) | AAV2/9-hEF1a-DIO-EYFP-WPRE-pA | Shanghai Taitool Bioscience Co. | Cat#S0196-9 | Viral titers:1.0x1012 particles/ml |
| Strain, strain background (AAV2/9) | AAV2/9-CAG-DIO-hGtACR1-P2A-EGFP-WPRE-pA | Shanghai Taitool Bioscience Co. | Cat# S0311-9 | Viral titers:5×1013 particles/ml |
| Strain, strain background (AAV2/9) | rAAV2/9-hSyn-DIO-mGFP-2A-Synaptophysin-mRuby | Shanghai Taitool Bioscience Co. | Cat# S0250-9 | Viral titers:1.55x1013 particles/ml |
| Strain, strain background (AAV2/2) | rAAV2/2-Retro-hEF1a-DIO-EYFP-WPRE-pA | Shanghai Taitool Bioscience Co. | Cat# S0196-2R | Viral titers: 2.52x1013 particles/ml |
| Strain, strain background (AAV2/9) | AAV2/9-hSyn-mCherry-WPRE-pA | Shanghai Taitool Bioscience Co. | Cat# S0238-9 | Viral titers:≥1.0 × 1013 particles/ml |
| Strain, strain background (AAV2/9) | AAV2/9-hSyn-hM4D(Gi)-mCherry-WPRE-pA | Shanghai Taitool Bioscience Co. | Cat# S0279-9 | Viral titers:≥1.0 × 1013 particles/ml |
| Strain, strain background (AAV2/9) | AAV2/9-hsyn-DIO-jGCaMP7s-WPRE-pA | Shanghai Taitool Bioscience Co. | Cat# S0590-9 | Viral titers:≥1.0 × 1013 particles/ml |
| Software, algorithm | ANY-Maze software 5.3 | Global Biotech Inc | http://www.anymaze.co.uk/ | |
| Software, algorithm | Image J | NIH | https://imagej.nih.gov/ij/index.html;%20 RRID:SCR_003070 | |
| Software, algorithm | GraphPad Prism 6 | GraphPad Software | https://www.graphpad.com/scientificsoftware/prism/; RRID: SCR_002798 | |
| Software, algorithm | MatLab R2016a | MathWorks | https://www.mathworks.com/products.html; RRID:SCR_001622 |
Numbers of retrogradely traced cells in CCK-Cre mice.
| Animal 1 | ||||||
|---|---|---|---|---|---|---|
| Region | Slice 1 | Slice 2 | Slice 3 | total | percentage | average |
| CC | 94 | 103 | 95 | 292 | 25.75% | 97 |
| DI | 11 | 34 | 21 | 66 | 5.82% | 22 |
| LPB | 71 | 103 | 69 | 243 | 21.43% | 81 |
| PAG | 26 | 34 | 19 | 79 | 6.97% | 26 |
| MO | 137 | 175 | 142 | 454 | 10.04% | 151 |
| Animal 2 | ||||||
| Region | Slice 1 | Slice 2 | Slice 3 | total | percentage | average |
| CC | 82 | 93 | 75 | 250 | 21.82% | 83 |
| DI | 18 | 23 | 17 | 58 | 5.06% | 19 |
| LPB | 94 | 102 | 91 | 287 | 25.04% | 96 |
| PAG | 11 | 23 | 15 | 49 | 4.28% | 16 |
| MO | 166 | 172 | 164 | 502 | 43.80% | 167 |
| Animal 3 | ||||||
| Region | Slice 1 | Slice 2 | Slice 3 | Total | percentage | average |
| CC | 80 | 95 | 76 | 251 | 24.25% | 84 |
| DI | 27 | 32 | 28 | 87 | 8.41% | 29 |
| LPB | 67 | 82 | 63 | 212 | 20.48% | 71 |
| PAG | 12 | 19 | 16 | 47 | 4.54% | 16 |
| MO | 139 | 157 | 142 | 438 | 42.32% | 146 |
| Region | Total average | |||||
| CC | 88 | |||||
| DI | 23 | |||||
| LPB | 83 | |||||
| PAG | 19 | |||||
| MO | 155 |
Data of behavioral tests after inhibition of LPBCCK neurons.
| Animal | Stage | Hot zone : time | Safe zone : latency to first entry | Hot zone time/total time | Optical fiber location | |
|---|---|---|---|---|---|---|
| GtACR1 | 1 | Habitat stage | 96.5 | Optical fibers in only one side | ||
| Test stage | 36.7 | 13.5 | 12.23% | |||
| 2 | Habitat stage | 60.8 | Optical fibers in only one side | |||
| Test stage | 70 | 67.0 | 23.33% | |||
| 3 | Habitat stage | 13.4 | ||||
| Test stage | 265.7 | 265.7 | 88.57% | |||
| 4 | Habitat stage | 151.2 | ||||
| Test stage | 300 | 300.0 | 100.00% | |||
| 5 | Habitat stage | 176.7 | ||||
| Test stage | 300 | 300.0 | 100.00% | |||
| 6 | Habitat stage | 70.6 | Optical fibers in only one side | |||
| Test stage | 93 | 91.7 | 31.00% | |||
| 7 | Habitat stage | 33.7 | ||||
| Test stage | 137.9 | 106.7 | 45.97% | |||
| EYFP | 8 | Habitat stage | 84.8 | |||
| Test stage | 72.9 | 2.3 | 24.30% | |||
| 9 | Habitat stage | 41.9 | ||||
| Test stage | 18.4 | 5.6 | 6.13% | |||
| 10 | Habitat stage | 14.9 | ||||
| Test stage | 6.7 | 2.4 | 2.23% | |||
| 11 | Habitat stage | 77.6 | ||||
| Test stage | 44.9 | 9.0 | 14.97% | |||
| 12 | Habitat stage | 137.3 | ||||
| Test stage | 52 | 7.5 | 17.33% | |||
| 13 | Habitat stage | 75.8 | ||||
| Test stage | 41.3 | 38.0 | 13.77% | |||
| 14 | Habitat stage | 149.4 | ||||
| Test stage | 108.9 | 108.9 | 36.30% | |||
| 15 | Habitat stage | 30.8 | ||||
| Test stage | 6.8 | 4.1 | 2.27% |
Number of labeled cells.
| Number of labeled cells | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| animal 1 | animal 2 | animal 3 | total average | ||||||||||||||||||
| region | slice 1 | slice 2 | slice 3 | total | percentage | average | region | slice 1 | slice 2 | slice 3 | total | percentage | average | region | slice 1 | slice 2 | slice 3 | total | percentage | average | |
| CC | 94 | 103 | 95 | 292 | 25.75% | 97 | CC | 82 | 93 | 75 | 250 | 21.82% | 83 | CC | 80 | 95 | 76 | 251 | 24.25% | 84 | 88 |
| DI | 11 | 34 | 21 | 66 | 5.82% | 22 | DI | 18 | 23 | 17 | 58 | 5.06% | 19 | DI | 27 | 32 | 28 | 87 | 8.41% | 29 | 23 |
| LPB | 71 | 103 | 69 | 243 | 21.43% | 81 | LPB | 94 | 102 | 91 | 287 | 25.04% | 96 | LPB | 67 | 82 | 63 | 212 | 20.48% | 71 | 83 |
| PAG | 26 | 34 | 19 | 79 | 6.97% | 26 | PAG | 11 | 23 | 15 | 49 | 4.28% | 16 | PAG | 12 | 19 | 16 | 47 | 4.54% | 16 | 19 |
| MO | 137 | 175 | 142 | 454 | 40.04% | 151 | MO | 166 | 172 | 164 | 502 | 43.80% | 167 | MO | 139 | 157 | 142 | 438 | 42.32% | 146 | 155 |