Pooled genome-wide CRISPR activation screening for rapamycin resistance genes in Drosophila cells
Figures
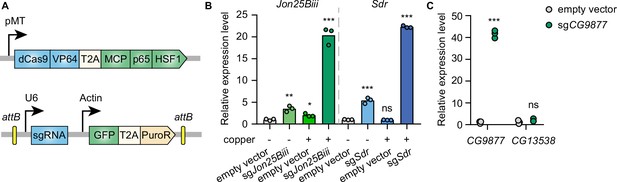
Inducible transcriptional activation by the synergistic activation mediator (SAM) complex in Drosophila cells.
(A) Schematic of the SAM complex for inducible transcriptional activation. dCas9-VP64 and MCP-p65-HSF1 were driven by an inducible metallothionein promoter. dCas9-VP64 and MCP-p65-HSF1 were expressed as T2A-containing bicistronic transcript. single-guide RNA (sgRNA) was expressed from pLib8 plasmid, which contains an attB flanking GFP-T2A-PuroR cassette for attP sites recombination. (B) Fold activation of Jon25Biii and Sdr expression measured by qPCR. Three biological replicates are shown as individual circles. (C) Fold activation of CG9877 and CG13538 expression measured by qPCR. Three biological replicates are shown as individual circles. t-test, *p<0.05; **p<0.01; ***p<0.001; ns, not significant.
-
Figure 1—source data 1
Full source data for Figure 1.
- https://cdn.elifesciences.org/articles/85542/elife-85542-fig1-data1-v1.zip
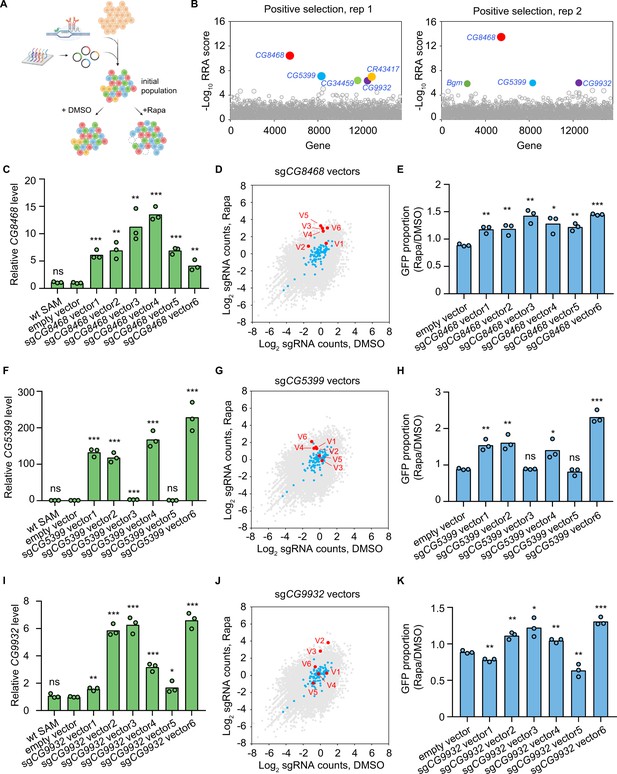
Genome-wide CRISPR activation screen for rapamycin resistance genes.
(A) Schematic of CRISPR activation screen (See methods). (B) Two replicates of genome-wide CRISPR activation screen. Data were analyzed by MAGeCK-RRA, a smaller RRA score indicates a stronger selection effect. Each circle represents a gene. Circle size corresponds to the significance (p value) of enrichment. Significantly enriched genes (false-discovery rate (FDR)<0.05) are colored. (C) Fold activation of CG8468 expression measured by qPCR. Three biological replicates are shown as individual circles. (D) Counts of sgCG8468 vectors from the genome-wide screen. Each dot represents a vector. Vectors targeting intergenic regions are shown in blue. Vectors targeting CG8468 are shown in red and annotated as V1-V6. (E) sgCG8468-expressing cell proliferation in cell mixture following 1 nM rapamycin or DMSO treatment. GFP proportion was measured by flow cytometry. Three biological replicates are shown as individual circles. (F) Fold activation of CG5399 expression measured by qPCR. Three biological replicates are shown as individual circles. (G) Counts of sgCG5399 vectors from the genome-wide screen. Each dot represents a vector. Vectors targeting intergenic regions are shown in blue. Vectors targeting CG5399 are shown in red and annotated as V1-V6. (H) sgCG5399-expressing cell proliferation in cell mixture following 1 nM rapamycin or DMSO treatment. GFP proportion was measured by flow cytometry. Three biological replicates are shown as individual circles. (I) Fold activation of CG9932 expression measured by qPCR. Three biological replicates are shown as individual circles. (J) Counts of sgCG9932 vectors from the genome-wide screen. Each dot represents a vector. Vectors targeting intergenic regions are shown in blue. Vectors targeting CG9932 are shown in red and annotated as V1-V6. (K) sgCG9932-expressing cell proliferation in cell mixture following 1 nM rapamycin or DMSO treatment. GFP proportion was measured by flow cytometry. Three biological replicates are shown as individual circles. t-test, *p<0.05; **p<0.01; ***p<0.001; ns, not significant.
-
Figure 2—source data 1
Full source data for Figure 2.
- https://cdn.elifesciences.org/articles/85542/elife-85542-fig2-data1-v1.zip
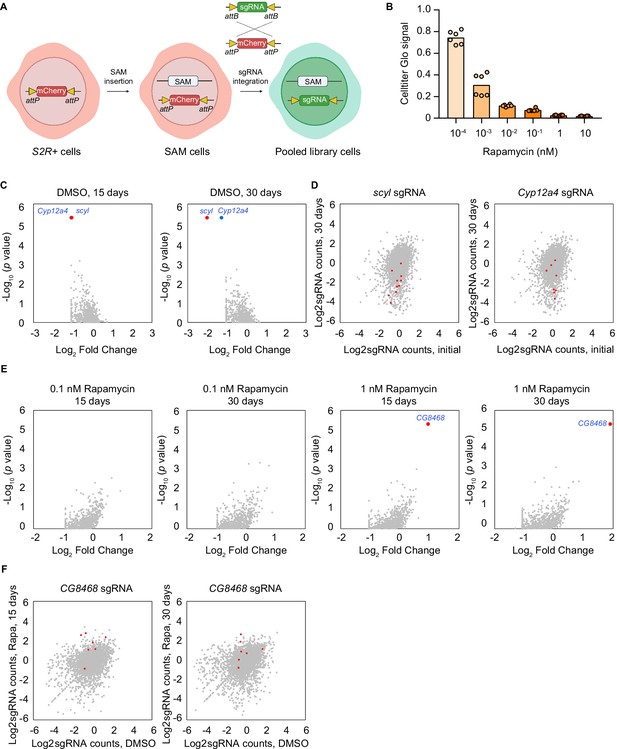
Pooled CRISPR activation screen with a focused library.
(A) Schematic of pooled library cells generation. Synergistic activation mediator (SAM) complex was inserted into attP sites containing S2R+ cells to generate SAM cells. The pooled attB sites flanking single-guide RNA (sgRNA) library was integrated into attP sites by phiC31-mediated recombination to generate pooled library cells. (B) Cell proliferation of S2R+ cells with rapamycin treatment at different concentrations. Cell proliferation was measured by Cell titer Glo. Six replicates are shown as individual circles. (C) Pooled focused library screen with DMSO treatment for 15 days and 30 days. Data were analyzed by MAGeCK. Each dot represents a gene. Genes with significant differences (false-discovery rate (FDR)<0.05) are highlighted. (D) Counts of sgRNAs targeting scyl or Cyp12a4 in the initial population and final population after proliferation for 30 days in DMSO. Each dot represents a sgRNA. sgRNAs targeting scyl or Cyp12a4 are highlighted. (E) Pooled-focused library screens with 0.1 nM and 1 nM rapamycin treatment for 15 days and 30 days. Data were analyzed by MAGeCK. Each dot represents a gene. Gene with a significant difference (FDR<0.05) is highlighted. (F) Counts of sgRNAs targeting CG8468 in the DMSO-treated population and rapamycin-treated population. Each dot represents a sgRNA. sgRNAs targeting CG8468 are highlighted.
-
Figure 2—figure supplement 1—source data 1
Full source data for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/85542/elife-85542-fig2-figsupp1-data1-v1.zip
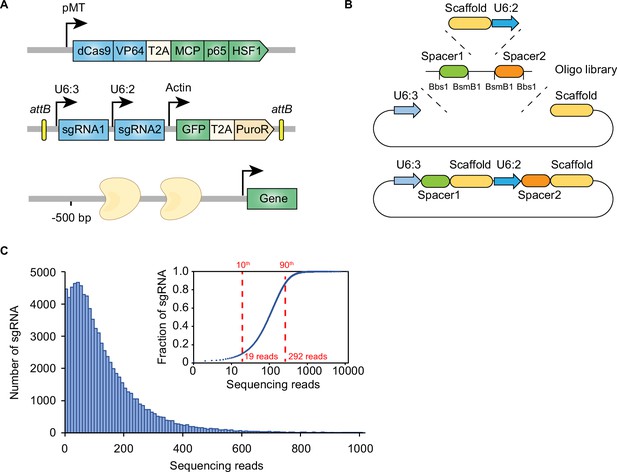
Design of genome-wide dual-sgRNA library.
(A) Schematic of dual-sgRNA library design. Two different sgRNAs targeting the same gene promoter region within 500 bp upstream of the transcriptional start site (TSS). Two sgRNAs were driven by separate U6 promoters. The attB sites flanking sgRNA sequence was integrated into attP sites flanking landing cassette by recombination-mediated cassette exchange (RMCE). (B) Schematic of genome-wide dual-sgRNA library construction by three-steps pooled cloning strategy. The oligo library containing Bbs1 and BsmB1 flanking sgRNA seed sequences was synthesized. The first library was constructed by inserting PCR products amplified from the oligo library. The second library was constructed by inserting the scaffold of the first sgRNA and the promoter of the second sgRNA using BsmB1 digestion and T4 ligation. The final library was constructed by inserting the sgRNA cassette into the destination vector using Bbs1 digestion and T4 ligation. (C) Histogram of sgRNA representation of the constructed dual-sgRNA library. Insert: Cumulative distribution of sgRNA sequencing reads. 10th and 90th percentiles are indicated by dash lines.
-
Figure 2—figure supplement 2—source data 1
Full source data for Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/85542/elife-85542-fig2-figsupp2-data1-v1.zip
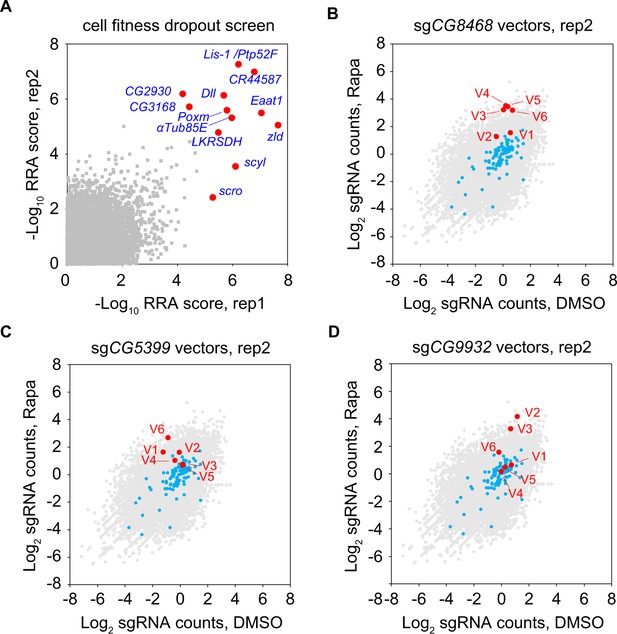
Genome-wide cell fitness screen and rapamycin screen.
(A) Genome-wide cell fitness screen. Each dot represents a gene. Significantly depleted genes (false-discovery rate (FDR)<0.05) after proliferation in DMSO for 3 weeks are highlighted. (B) Counts of sgCG8468 vectors in genome-wide rapamycin screen. Each dot represents a vector. Vectors targeting the intergenic regions are shown in blue. Vectors targeting the candidates are shown in red and annotated as V1-V6. (C) Counts of sgCG5399 vectors in genome-wide rapamycin screen. (D) Counts of sgCG9932 vectors in genome-wide rapamycin screen.
-
Figure 2—figure supplement 3—source data 1
Full source data for Figure 2—figure supplement 3.
- https://cdn.elifesciences.org/articles/85542/elife-85542-fig2-figsupp3-data1-v1.zip
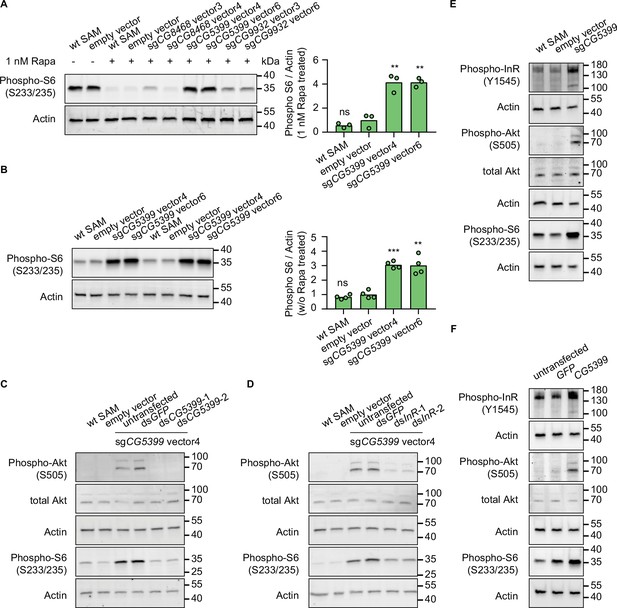
CG5399 overexpression activates RTK-Akt-mTOR signaling.
(A) Phospho-S6 levels in cells expressing dual-sgRNA vectors in the presence of 1 nM rapamycin. Western blot signals are quantitatively analyzed by ImageJ. Three biological replicates are shown as individual circles. (B) Phospho-S6 levels in cells expressing sgCG5399 vectors without rapamycin treatment. Western blot signals are quantitatively analyzed by ImageJ. Four biological replicates are shown as individual circles. (C) Phospho-Akt and Phospho-S6 in CG5399-overexpressing cells following CG5399 knockdown. Two nonoverlapping double-stranded RNAs (dsRNAs) targeting CG5399 were used. (D) Phospho-Akt and Phospho-S6 in CG5399-overexpressing cells following insulin receptor (InR) knockdown. Two nonoverlapping dsRNAs targeting InR were used. (E) Phospho-InR, phospho-Akt, and phospho-S6 in sgCG5399-expressing synergistic activation mediator (SAM) cells. (F) Phospho-InR, phospho-Akt, and phospho-S6 in CG5399 ORF-overexpressing S2R+ cells using UAS-Gal4. t-test, **<0.01; ***p<0.001; ns, not significant.
-
Figure 3—source data 1
Full source data for Figure 3.
- https://cdn.elifesciences.org/articles/85542/elife-85542-fig3-data1-v1.zip
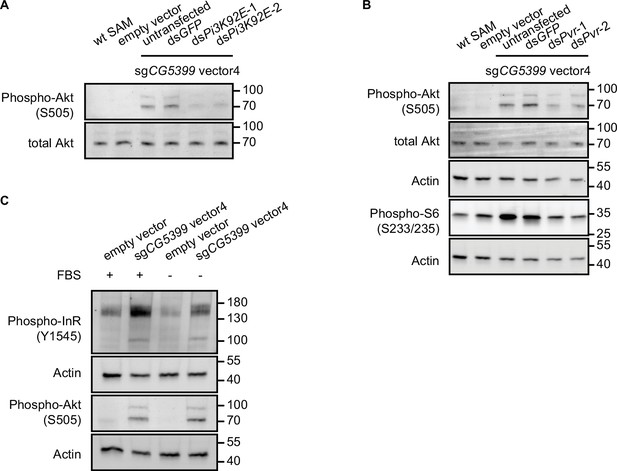
CG5399 overexpression activates Akt-mTOR through RTK/PI3K.
(A) Phospho-Akt in CG5399-overexpressing cells following Pi3K92E knockdown. Two nonoverlapping double-stranded RNAs (dsRNAs) targeting Pi3K92E were used. (B) Phospho-Akt and phospho-S6 in CG5399-overexpressing cells following PDGF/VEGF receptor (Pvr) knockdown. Two nonoverlapping dsRNAs targeting Pvr were used. (C) Phospho-InR and phospho-Akt in CG5399-overexpressing cells following serum starvation for 2 hr.
-
Figure 3—figure supplement 1—source data 1
Full source data for Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/85542/elife-85542-fig3-figsupp1-data1-v1.zip
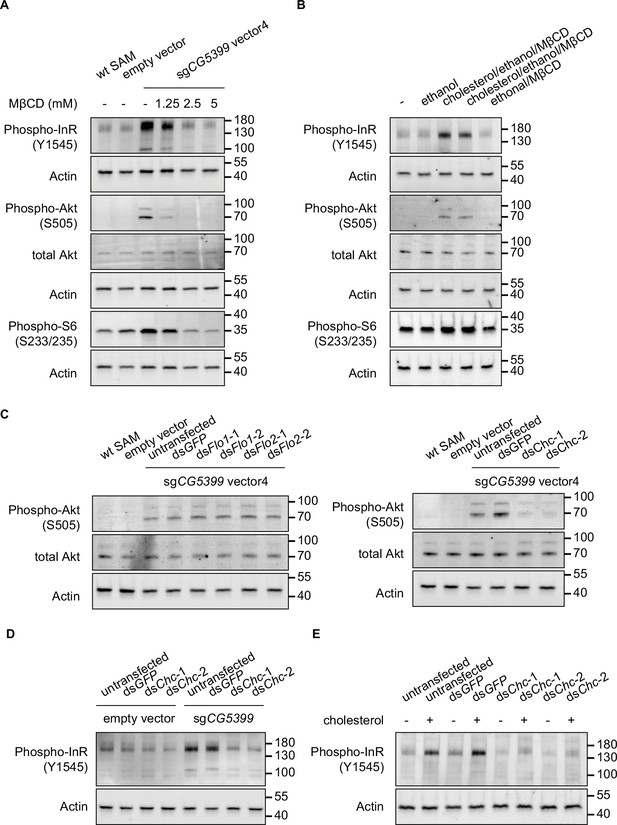
Activation of InR-Akt-mTOR signaling by CG5399 overexpression requires cholesterol and clathrin-coated pits at the membrane.
(A) Phospho-InR, phospho-Akt, and phospho-S6 in CG5399-overexpressing cells treated with methyl-beta-cyclodextrin (MβCD) at different concentrations. (B) Phospho-InR, phospho-Akt, and phospho-S6 in S2R+ cells with cholesterol supplementation. Two different cholesterol products from Sigma (C3045 for Lane 3 and C2044 for Lane 4) were used. (C) Phospho-Akt in CG5399-overexpressing cells following Flo1, Flo2, or Chc knockdown. Two nonoverlapping double-stranded RNAs (dsRNAs) targeting each gene were used. (D) Phospho-InR in CG5399-overexpressing cells following Chc knockdown. Two nonoverlapping dsRNAs targeting each gene were used. (E) Phospho-InR in S2R+ cells with cholesterol supplementation following clathrin heavy chain (Chc) knockdown. Two nonoverlapping dsRNAs targeting each gene were used.
-
Figure 4—source data 1
Full source data for Figure 4.
- https://cdn.elifesciences.org/articles/85542/elife-85542-fig4-data1-v1.zip
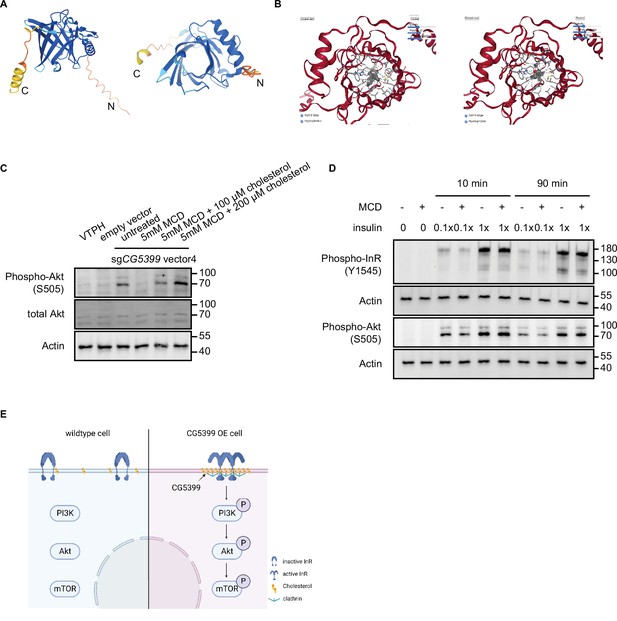
CG5399 interacts with cholesterol by molecular docking.
(A) Structure prediction of CG5399 by AlphaFold. Different side views are shown. (B) Interaction between CG5399 and cholesterol by molecular docking. Cholesterol is predicted to be inserted into the barrel structure of CG5399 in two different configurations. (C) Phospho-Akt in CG5399-overexpressing cells supplemented with cholesterol following methyl-beta-cyclodextrin (MβCD) treatment. (D) Phospho-InR, phospho-Akt in S2R+ cells stimulated with insulin following MβCD treatment. Different insulin concentrations and treatment durations were tested. (E) Schematic model of InR-Akt-mTOR activation in CG5399 overexpressing cells.
-
Figure 4—figure supplement 1—source data 1
Full source data for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/85542/elife-85542-fig4-figsupp1-data1-v1.zip
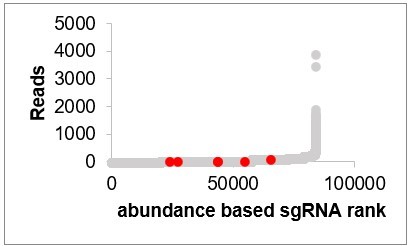
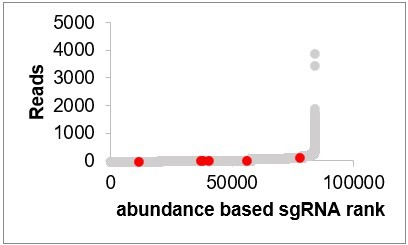
Reads of CG5399 sgRNAs in the initial population (CG5399 sgRNAs are in red, other sgRNAs are in gray).

Reads of CG8468 sgRNAs in the initial population (CG8468 sgRNAs are in red, other sgRNAs are in gray).
Tables
Significantly depleted genes in genome-wide fitness screen.
| Gene | Human ortholog | Known gene affecting cell fitness | Reference |
|---|---|---|---|
| zld | ZNF485 | ||
| Eaat1 | SLC1A3 | ||
| CR44587 | - | ||
| Lis-1/Ptp52F* | PAFAH1B1/Ptprb | LIS-1-overexpressing mitotic cells show a variety of spindle defects | PMID: 10722879 |
| scyl | DDIT4 | Scyl inhibits cell growth by regulating the Tor pathway | PMID: 15545626 |
| αTub85E | TUBA1A | ||
| Poxm | Pax9 | PAX9 overexpression inhibits cancer cell proliferation | PMID: 35628401 |
| Dll | DLX6 | ||
| LKRSDH | AASS | Overexpression of Aass suppresses cancer cell proliferation | PMID: 31601242 |
| scro | NKX2-1 | NKX2-1 suppresses lung cancer progression by dampening ERK activity | PMID: 34689179 |
| CG3168 | SV2A | Overexpression of SV2A inhibits the PI3K signaling pathway | PMID: 34277597 |
| CG2930 | SLC15A1 |
-
*
Lis-1 and Ptp52F form divergent gene pair ~500 bp apart.
Overlapping genes of top-ranked 50 hits from two genome-wide screen replicates.
| Rank (Rep 1, Rep 2) | Gene | Human ortholog | Function | Known rapamycin resistance gene |
|---|---|---|---|---|
| 1, 1 | CG8468 | SLC16A8 | monocarboxylate transporter | |
| 2, 3 | CG5399 | APOD/LCN2 | lipocalin | |
| 5, 2 | CG9932 | ZFN462/REST | transcription factor | |
| 4, 10 | CG34459 | / | unknown | |
| 22, 13 | Pka-C3 | PRKX | catalytic subunit of PKA | PMID: 15643061, 14673167, 11739804 |
| 41, 8 | Ps | NOVA1 | RNA splicing | |
| 43, 45 | CDC25 | CDC25A/CDC25B | tyrosine phosphatase | PMID: 24383842, 19276368 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | CG8468 | FlyBase | FLYB:FBgn0033913 | |
| Gene (Drosophila melanogaster) | CG5399 | FlyBase | FLYB:FBgn0038353 | |
| Gene (Drosophila melanogaster) | CG9932 | FlyBase | FLYB:FBgn0262160 | |
| Cell line (D. melanogaster) | S2R+ | DRSC | FLYB:FBtc0000150 | |
| Cell line (D. melanogaster) | PT5 | DRSC | FLYB:FBtc0000229 | |
| Strain, strain background (Escherichia coli) | E.cloni10GF’ Electrocompetent Cells | Biosearch Technologies | 60061–2 | sgRNA library construction |
| Strain, strain background (Escherichia coli) | One Shot TOP10 Chemically Competent E. coli | Invitrogen | C404010 | |
| Antibody | Recombinant Anti-Insulin Receptor (phospho Y1185) antibody (Rabbit monoclonal) | Abcam | ab62321 | 1:1000 for WB |
| Antibody | Phospho-Akt (Ser473) (D9E) XP antibody (Rabbit monoclonal) | Cell Signaling Technology | 4060 | 1:1000 for WB |
| Antibody | Akt Rabbit Antibody (Rabbit polyclonal) | Cell Signaling Technology | 9272 | 1:1000 for WB |
| Antibody | StarBright Blue 700 Goat Anti-Rabbit IgG | Bio-Rad | 12004161 | 1:2500 for WB |
| Antibody | StarBright Blue 520 Goat Anti-Rabbit IgG | Bio-Rad | 12005869 | 1:2500 for WB |
| Antibody | hFAB Rhodamine Anti-Actin Primary Antibody (synthesized, monoclonal) | Bio-Rad | 12004163 | 1:2500 for WB |
| Recombinant DNA reagent | pMK33-SAM plasmid | This paper | Can be obtained from DRSC | |
| Recombinant DNA reagent | pLib8 plasmid | This paper | U6:3-MS2 sgRNA cassette, can be obtained from DRSC | |
| Recombinant DNA reagent | pBS130 plasmid | Addgene | 26290 | PhiC31 integrase |
| Recombinant DNA reagent | pUAS-CG5399 plasmid | This paper | CG5399 ORF vector, cassette, can be obtained from DRSC | |
| Commercial assay or kit | Effectene Transfection Reagent | Qiagen | 301425 | |
| Commercial assay or kit | CellTiter-Glo Luminescent Cell Viability Assay | Promega | G7570 | |
| Commercial assay or kit | RNeasy Mini Kit | Qiagen | 74104 | |
| Commercial assay or kit | iScript cDNA Synthesis Kit | Bio-Rad | 1708890 | |
| Chemical compound, drug | MEGAscript T7 Transcription Kit | Invitrogen | AM1334 | |
| Chemical compound, drug | Methyl-β-cyclodextrin | Sigma-Aldrich | C4555 | |
| Chemical compound, drug | Cholesterol | Sigma-Aldrich | C3045 | |
| Chemical compound, drug | Cholesterol | Sigma-Aldrich | C2044 | |
| Software, algorithm | GraphPad Prism 7 | GraphPad | ||
| Software, algorithm | FlowJo | FlowJo |
Additional files
-
Supplementary file 1
sgRNA vectors used in this study.
- https://cdn.elifesciences.org/articles/85542/elife-85542-supp1-v1.docx
-
Supplementary file 2
PCR primers used in this study.
- https://cdn.elifesciences.org/articles/85542/elife-85542-supp2-v1.docx
-
Supplementary file 3
dsRNAs used in this study.
- https://cdn.elifesciences.org/articles/85542/elife-85542-supp3-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85542/elife-85542-mdarchecklist1-v1.docx
-
Source data 1
The original files of the full raw unedited blots and figures with the uncropped blots with relevant bands labeled in this study.
- https://cdn.elifesciences.org/articles/85542/elife-85542-data1-v1.zip