Multimodal HLA-I genotype regulation by human cytomegalovirus US10 and resulting surface patterning
Figures
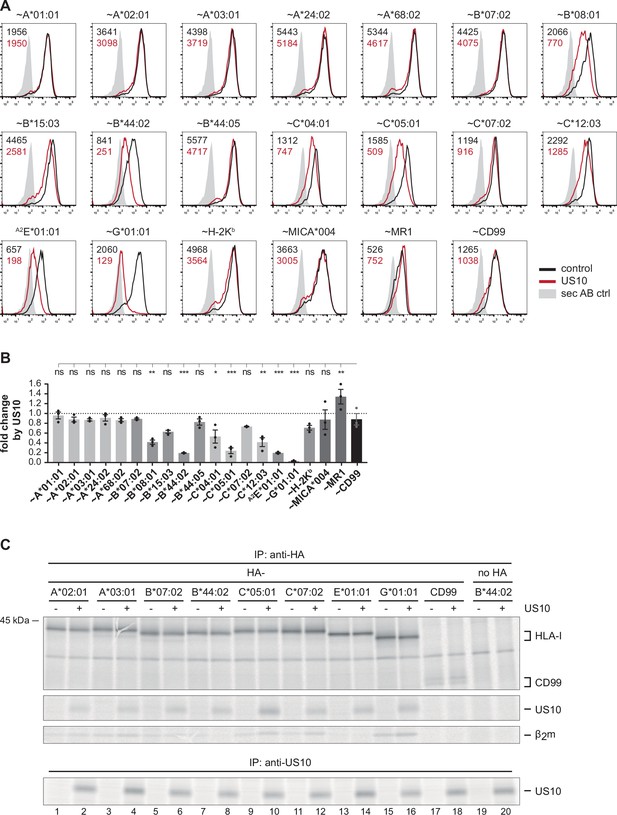
Geno- and allotype-specific regulation of HLA-I by US10.
(A) HeLa cells were transiently co-transfected with plasmids encoding HA-tagged (~) molecules or non-tagged HLA-A2E*01:01 (HLA-E was expressed with an HLA-A*02 signal peptide, a natural HLA-E ligand) and plasmids with either US10 or a control protein together with an IRES-EGFP cassette. Surface expression was determined by flow cytometry (anti-HA or anti-HLA-E) on EGFP-positive cells. Representative histograms are shown. (B) Fold change of surface expression by US10 was calculated as the ratio of the median fluorescence intensity (MFI) of US10-expressing cells compared to control transfected cells. Dots represent individual values and bars mean values ± SEM from three independent experiments (biological replicates). Significance compared to the HA-CD99 control was calculated using one-way ANOVA followed by Dunnett’s multiple comparison test. (C) HeLa cells were transiently transfected as described in (A) and metabolically labeled for 2 hr. Digitonin cell lysates were prepared, and immunoprecipitations using anti-HA or anti-US10 were performed and separated by SDS-PAGE with subsequent detection by autoradiography (Figure 1—source data 1). One of two independent experiments is shown.
-
Figure 1—source data 1
Immunoprecipitation in Figure 1C.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig1-data1-v1.zip
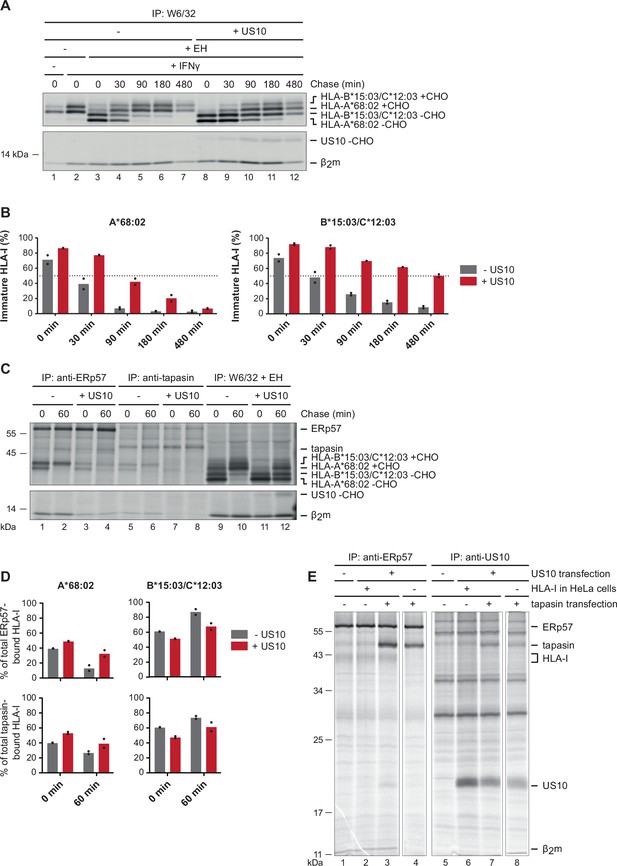
US10 blocks human leucocyte antigen class I (HLA-I) interaction with the peptide loading complex (PLC).
(A) Control HeLa cells or cells stably expressing US10 were induced by IFNγ overnight and subsequently metabolically labeled for 30 min and chased as indicated. After immunoprecipitation by W6/32, proteins were digested by EndoH (-CHO, deglycosylated proteins; +CHO, resistant glycans) as indicated and separated by SDS-PAGE. Labeled proteins were detected by autoradiography (Figure 2—source data 1). (B) The intensities of single HLA-I heavy chain (HC) bands in (A) were quantified, and the percentage of immature molecules compared to the total amount (sum of immature and mature) was calculated and depicted from two independent experiments (biological replicates). (C) Immunoprecipitation from HeLa cells or cells stably expressing US10 was performed as in (A) but with modified chase times and without IFNγ treatment. Antibodies applied for immunoprecipitations are indicated (Figure 2—source data 2). (D) Band intensities of HLA-I HCs in anti-ERp57 and anti-tapasin immunoprecipitations from (C) were quantified and the amount of the HLA-A*68:02 HC (left panel) and HLA-B*15:03/-C*12:03 HC (right panel) was calculated as the percentage of total PLC-bound HLA-I (sum of both HC bands). Dots represent individual values from two independent experiments (biological replicates). (E) Wild-type or HLA-I KO HeLa cells were transiently transfected with US10 and tapasin-expressing plasmids as indicated. At 20 hr post-transfection, cells were metabolically labeled for 3 hr. Immunoprecipitation was performed with anti-ERp57 or anti-US10 antibodies (Figure 2—source data 3). One of two independent experiments is shown in panels (A), (C), and (E).
-
Figure 2—source data 1
Immunoprecipitation in Figure 2A.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig2-data1-v1.zip
-
Figure 2—source data 2
Immunoprecipitation in Figure 2C.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig2-data2-v1.zip
-
Figure 2—source data 3
Immunoprecipitation in Figure 2E.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig2-data3-v1.zip
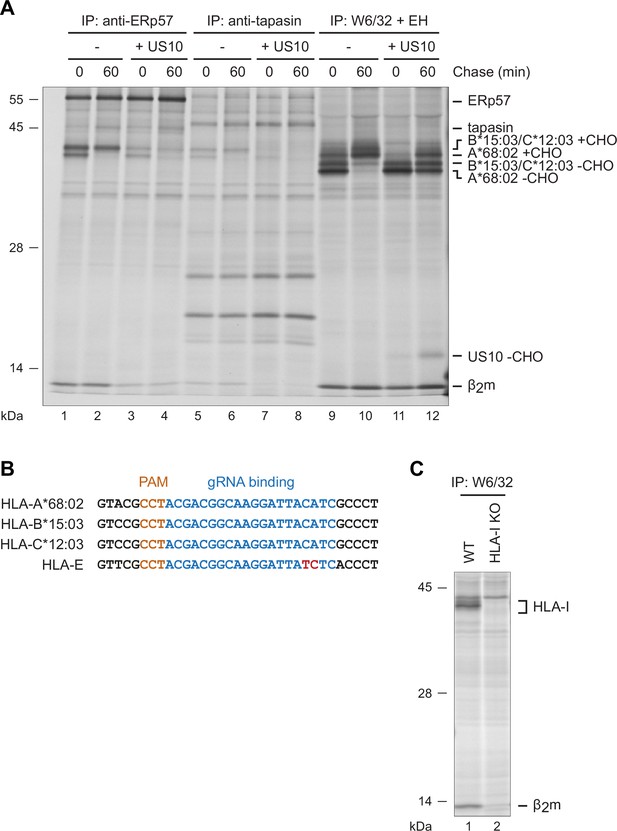
US10 blocks human leucocyte antigen class I (HLA-I) interaction with the peptide loading complex (PLC).
(A) Experiment from Figure 2C showing the full size of the gel (Figure 2—figure supplement 1—source data 1). (B) Alignment of the sequences of HeLa HLA-I (HLA-A*68:02, -B*15:03, -C*12:03 and -E). Highlighted are the PAM (orange) and the gRNA binding (blue) sites used for generation of the HLA-I knockout HeLa cells. A mismatch in the HLA-E sequence is indicated in red. (C) Wild-type and HLA-I KO HeLa cells were metabolically labeled for 2 hr prior to immunoprecipitation using W6/32. Retrieved proteins were separated by SDS-PAGE and detected by autoradiography.
-
Figure 2—figure supplement 1—source data 1
Immunoprecipitation in Figure 2—figure supplement 1A.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig2-figsupp1-data1-v1.zip
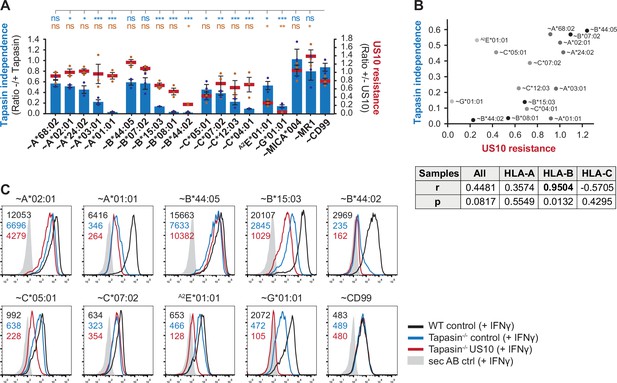
Higher tapasin dependency in HLA-B correlates with increased sensitivity to US10.
(A) Wild-type or tapasin knockout HeLa cells were transiently co-transfected as indicated and treated with IFNγ overnight. Cell surface expression of the HA-tagged molecules or non-tagged HLA-A2E*01:01 was determined (representative histograms in Figure 3—figure supplement 1B) as in Figure 1A. US10 resistance was calculated as the ratio of the median fluorescence intensity (MFI) of US10-expressing cells compared to control cells (red lines). Tapasin independence was calculated as the ratio of the MFI of tapasin knockout cells compared to wild-type cells (blue bars). Dots represent individual values and bars mean values ± SEM from three independent experiments (biological replicates). Significance compared to the HA-CD99 control was calculated using one-way ANOVA followed by Dunnett’s multiple comparison test. (B) Two-tailed correlation analysis of the results from (A). (C) Flow cytometry analysis performed as in (A) including US10 in tapasin knockout cells. Representative histograms from one of three independent experiments (biological replicates) are shown.
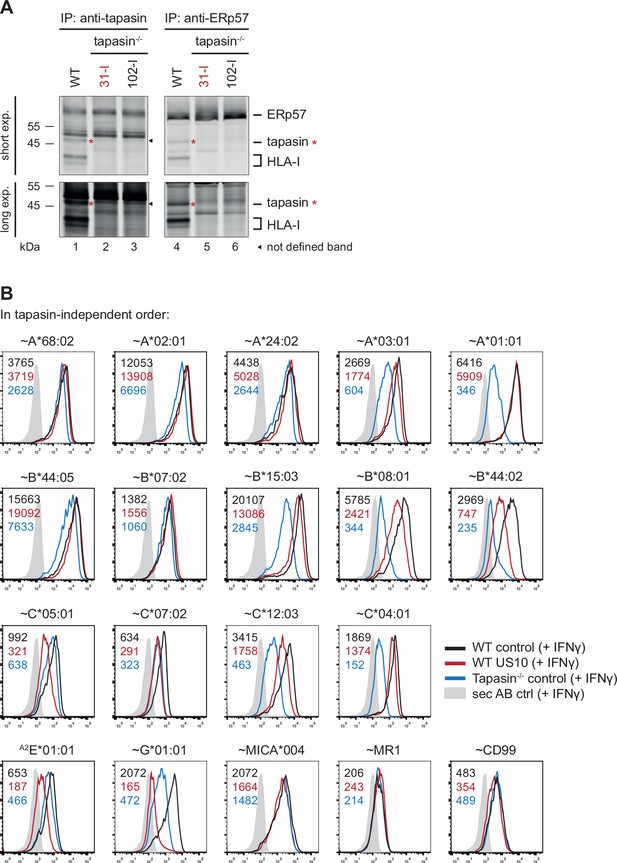
HLA-B regulation by US10 correlates with HLA-B tapasin dependency.
(A) Wild-type and two clones of tapasin KO HeLa cells (31-I and 102-I) were metabolically labeled for 2 hr. Immunoprecipitations using anti-tapasin and anti-ERp57 were performed. Red asterisk: tapasin. Black arrow: unspecific band. A short and a long exposure are shown (Figure 3—figure supplement 1—source data 1). Tapasin KO clone 31-I was used for all subsequent experiments. (B) Representative histograms for Figure 3A.
-
Figure 3—figure supplement 1—source data 1
Immunoprecipitation in Figure 3—figure supplement 1A.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig3-figsupp1-data1-v1.zip
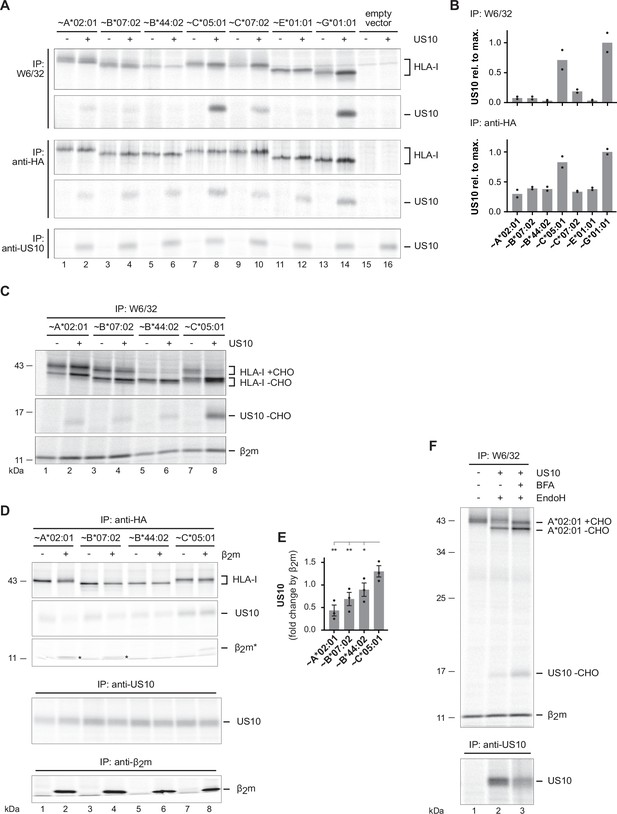
US10 binding to β2m/HC heterodimers correlates with human leucocyte antigen class I (HLA-I) endoplasmic reticulum (ER) retention.
(A) HLA-I KO HeLa cells were transiently co-transfected with indicated HA-HLA-I-expressing plasmids comprising a mutated gRNA binding site together with a US10- or a control-pIRES-EGFP plasmid. To improve assembly of HLA-E, UL40 (comprising an HLA-E ligand) was expressed with HLA-E. Cells were metabolically labeled for 2 hr and immunoprecipitations were performed as in Figure 1C; antibodies were applied as indicated on the left (Figure 4—source data 1 and 2). (B) Relative signal strengths from single bands of US10 in the W6/32 and (upper panel) anti-HA immunoprecipitation (lower panel) samples are shown. Dots represent individual values and bars mean values thereof from two independent experiments (biological replicates). The ratio (US10/control) of single bands of HLA-I HCs in the anti-HA and W6/32 immunoprecipitation samples is shown. Dots represent individual values and bars mean values thereof from two independent experiments (biological replicates). (C) HLA-I KO HeLa cells were transfected and treated as in (A). Immunoprecipitation was performed with W6/32 and subsequently an EndoH digest was performed (Figure 4—source data 3). (D) HLA-I/β2m, double KO HeLa cells were transiently transfected with US10, HA-tagged HLA-I, and β2m as indicated. At 20 hr post-transfection, cells were metabolically labeled for 2 hr and immunoprecipitation was performed as indicated (Figure 4—source data 4). One of three independent experiments is shown. (E) The intensity of the US10 bands co-immunoprecipitated with anti-HA was quantified, and the ratios of the samples with and without β2m were determined from three independent experiments (biological replicates). Significance was calculated using one-way paired ANOVA followed by Dunnett’s multiple comparison test. (F) HLA-I KO HeLa cells were transfected with HA-HLA-A*02:01 and US10 or a control plasmid. At 20 hr post-transfection, cells were treated with brefeldin A (BFA) during metabolic labeling for 2 hr. Subsequently, an immunoprecipitation using anti-HA was performed. Indicated samples were subjected to EndoH digestion prior to SDS-PAGE separation (Figure 4—source data 5). One of two independent experiments is shown in (A), (C), and (F).
-
Figure 4—source data 1
Immunoprecipitations with W6/32 and anti-HA in Figure 4A.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig4-data1-v1.zip
-
Figure 4—source data 2
Immunoprecipitation with anti-US10 in Figure 4A.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig4-data2-v1.zip
-
Figure 4—source data 3
Immunoprecipitation in Figure 4C.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig4-data3-v1.zip
-
Figure 4—source data 4
Immunoprecipitation in Figure 4D.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig4-data4-v1.zip
-
Figure 4—source data 5
Immunoprecipitation in Figure 4F.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig4-data5-v1.zip
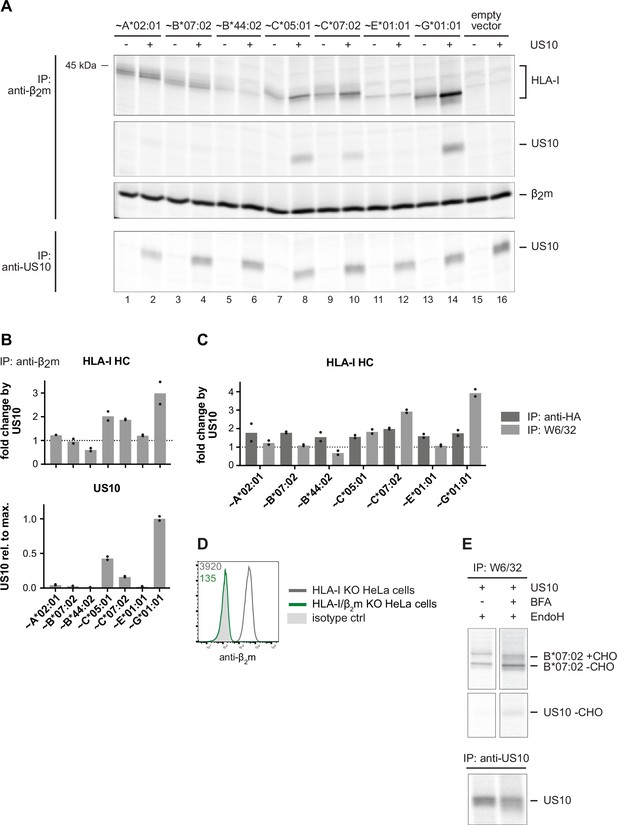
US10 forms stable complexes with assembled HLA-C and -G, but not with HLA-A and -B.
(A) Immunoprecipitations were performed as described in Figure 4A using the antibodies anti-β2m and anti-US10 (Figure 4—figure supplement 1—source data 1). (B) The ratio (US10/control) of single bands of human leucocyte antigen class I (HLA-I) heavy chain (HCs) (upper part) and relative signal strengths from US10 bands (lower part) in the anti-β2m immunoprecipitation samples are shown. Dots represent individual values and bars mean values thereof from two independent experiments (biological replicates). (C) The ratio (US10/control) of single bands of HLA-I HCs in the W6/32 and anti-HA immunoprecipitations from Figure 4A. Dots represent individual values and bars mean values thereof from two independent experiments (biological replicates). (D) Intracellular flow cytometry analysis of HLA-I/β2m double knockout HeLa cells. Staining with anti-β2m is shown. (E) Analysis of HA-HLA-B*07:02 using the conditions described in Figure 4F (Figure 4—figure supplement 1—source data 2).
-
Figure 4—figure supplement 1—source data 1
Immunoprecipitations in Figure 4—figure supplement 1A.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Immunoprecipitations in Figure 4—figure supplement 1E.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig4-figsupp1-data2-v1.zip
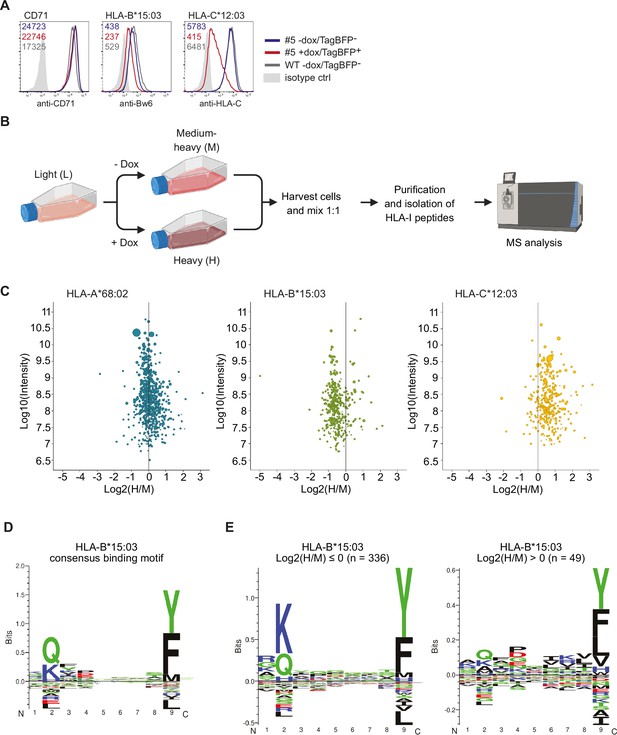
Quantitative human leucocyte antigen class I (HLA-I) ligandome analysis confirms genotype-dependent effects by US10.
(A) Wild-type HeLa cells or TagBFP-T2A-US10i clone #5 were treated with doxycycline (0.33 µg/mL) or DMSO for 24 hr. Subsequently. cells were stained by anti-CD71, anti-Bw6, or anti-HLA-C antibodies and a flow cytometry analysis was performed (cells were gated according to TagBFP expression, TagBFP+ or TagBFP-). (B) Experimental scheme of pSILAC immunopeptidomics. HeLa TagBFP-T2A-US10i #5 was pulse-labeled with medium-heavy (M) or heavy (H) amino acids in the presence of DMSO or doxycycline, respectively. After 24 hr, metabolically labeled cells were harvested and combined in a 1:1 ratio. HLA-I peptides were isolated and analyzed by nanoLC-MS/MS. The scheme was created with BioRender.com (C) Scatter plots showing median log2 H/M ratios and log10 intensity of quantified HLA-I peptides with respect to their HLA-I allele. Dot size correlates with number of ratios used to calculate the corresponding peptide ratio. (D) HLA-I consensus binding motif of HLA-B*15:03 obtained from NetMHCpan 4.1 motif viewer (https://services.healthtech.dtu.dk/services/NetMHCpan-4.1/) (E) Gibbs clustering analysis (GibbsCluster 2.0) for quantified HLA-B*15:03 peptides with median log2 H/M ratios ≤0 or >0.
© 2024, BioRender Inc. Figure 5 was created using BioRender, and is published under a CC BY-NC-ND 4.0. Further reproductions must adhere to the terms of this license.
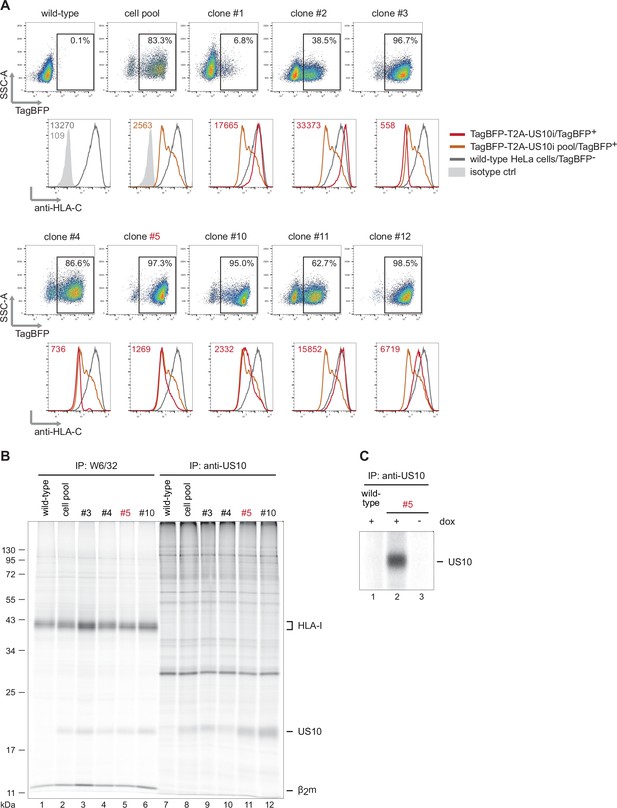
Quantitative human leucocyte antigen class I (HLA-I) ligandome analysis confirms genotype-dependent effects by US10.
(A, B) TagBFP-T2A-US10i HeLa cell pool and cell clones and wild-type HeLa cells were treated with doxycycline (0.33 µg/mL) for 24 hr. In (A), TagBFP expression was analyzed by flow cytometry (dot plots) and HLA-C expression (histograms) was determined on the TagBFP-positive population, except for wild-type HeLa cells, for which the TagBFP-negative population is shown. In (B), cells were metabolically labeled for 2 hr and immunoprecipitations were performed either with W6/32 or an anti-US10 antibody. Samples were separated by SDS-PAGE and detected by autoradiography (Figure 5—figure supplement 1—source data 1). (C) Wild-type HeLa cells or TagBFP-T2A-US10i cell clone #5 were treated with doxycycline (0.33 µg/mL) or DMSO for 24 hr. An immunoprecipitation was performed as in (B) with an anti-US10 antibody (Figure 5—figure supplement 1—source data 2).
-
Figure 5—figure supplement 1—source data 1
Immunoprecipitations in Figure 5—figure supplement 1B.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
Immunoprecipitations in Figure 5—figure supplement 1C.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig5-figsupp1-data2-v1.zip
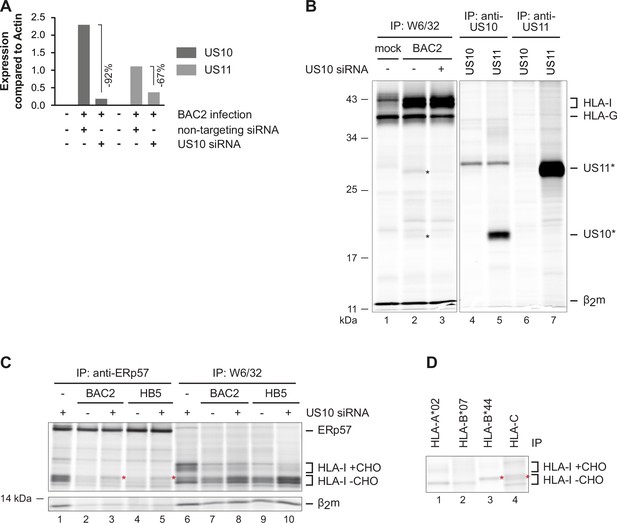
Downregulation of overlapping US10 and US11 transcripts in human cytomegalovirus (HCMV)-infected cells rescues human leucocyte antigen class I (HLA-I) interaction with the peptide loading complex (PLC).
(A) MRC-5 fibroblasts were nucleofected with US10-specific or non-targeting siRNA 24 hr prior to mock treatment or infection with HCMV ΔUS2-6 mutant BAC2 at an MOI (multiplicity of infection) of 5. At 24 hr p.i., RNA was isolated. Subsequently, cDNA was generated and analyzed by qPCR. The binding sites for the used primers are depicted in Figure 6—figure supplement 2. Expression of US10 and US11 is shown compared to expression of actin. (B) HLA-G-expressing BJ-5ta fibroblasts were treated with siRNA and infected as in (A). At 48 hr post-infection, cells were metabolically labeled for 2 hr. Digitonin cell lysates were prepared, and immunoprecipitations were performed as indicated (lanes 1–3). In parallel, immunoprecipitations were performed with HeLa cells transfected with US10 or US11 expression plasmids, lanes 4–7 (Figure 6—source data 1). (C) MRC-5 fibroblasts were nucleofected with US10-specific or non-targeting siRNA 24 hr prior to mock treatment or infection with the HCMV ΔUS2-6 mutants BAC2 or HB5 at an MOI of 7. Proteins were metabolically labeled at 24 h p.i. for 2 hr, and immunoprecipitations using anti-ERp57 or W6/32 were performed. All samples were treated by EndoH (Figure 6—source data 2). Asterisk: strongly increased HLA-I HC when applying US10 siRNA. One of two independent experiments (biological replicates) is shown. (D) BAC2-infected MRC-5 fibroblasts were treated as in (C), and HLA-I-specific immunoprecipitations were performed as indicated. All samples were EndoH-treated prior to separation by SDS-PAGE (Figure 6—source data 2).
-
Figure 6—source data 1
Immunoprecipitation in Figure 6B.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig6-data1-v1.zip
-
Figure 6—source data 2
Immunoprecipitations in Figure 6C and D.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig6-data2-v1.zip
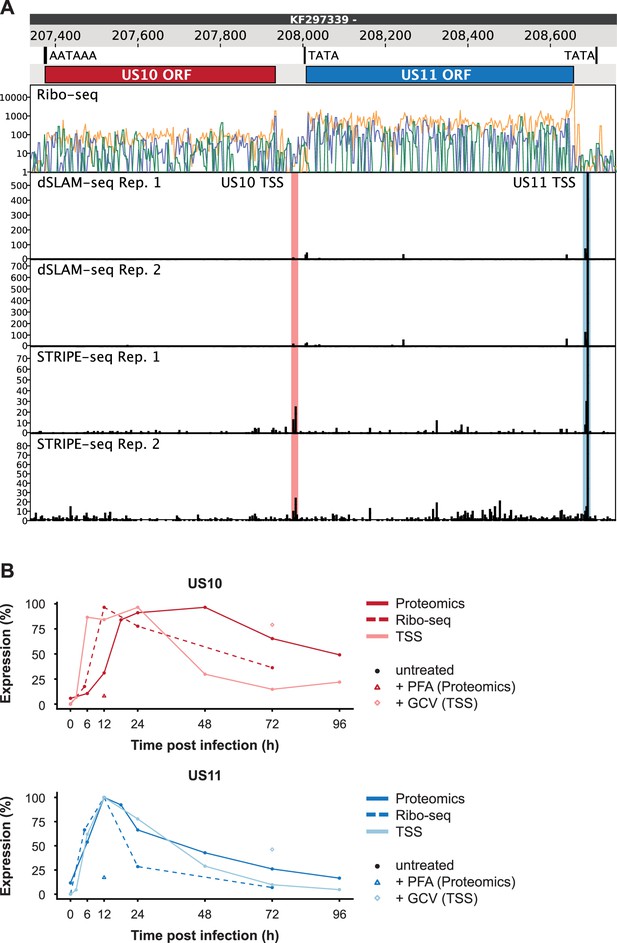
Overlapping US10 and US11 transcripts in human cytomegalovirus (HCMV)-infected cells.
(A) Genome browser showing the US10/11 locus. The tracks show (from top to bottom) the genomic position, identified sequence motifs, the known open-reading frames (ORFs) encoding US10 and US11, the Ribo-seq signal mapped to the three possible reading frames (orange, green, blue), and the 5’ end read counts for TSS profiling data. Identified US10 and US11 transcription start sites (TSS) are indicated. (B) Time-course plots depicting the temporal expression dynamics of US10 (top) and US11 (bottom). TSS profiling, Ribo-seq, and proteomics data were scaled to the corresponding maximal value. Samples that were treated with genome replication inhibitors (PFA or GCV) are indicated.
-
Figure 6—figure supplement 1—source data 1
Western blots in Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig6-figsupp1-data1-v1.zip
-
Figure 6—figure supplement 1—source data 2
Immunoprecipitation in Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig6-figsupp1-data2-v1.zip
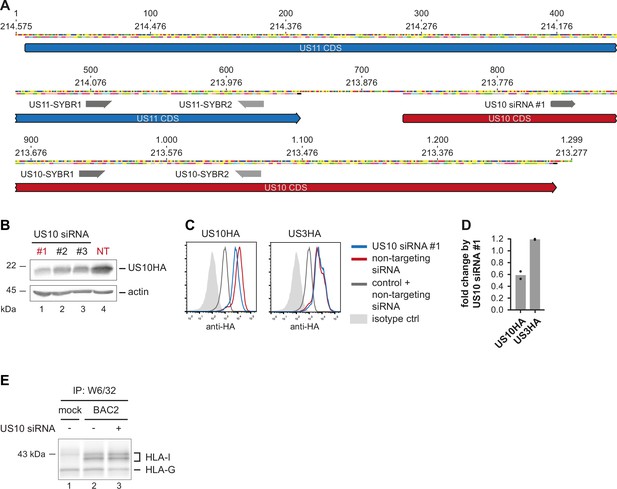
Downregulation of overlapping US10 and US11 transcripts in human cytomegalovirus (HCMV)-infected cells rescues human leucocyte antigen class I (HLA-I) interaction with the peptide loading complex (PLC).
(A) Depiction of the coding sequences (CDS) for US11 and US10 and binding sites for the primers used for RT-PCR in Figure 6A and of US10-targeting siRNA #1. (B) Stable HeLa-US10HA cells were transfected with three different siRNA targeting US10 (#1–3) or with non-targeting (NT) siRNA. Whole-cell lysates were prepared for western blot analysis using anti-HA antibodies (Figure 6—figure supplement 2—source data 1). US10-targeting siRNA #1 was used for all subsequent experiments. (C, D) MRC-5 fibroblasts were nucleofected with US10HA, US3HA, or a control plasmid together with US10- or non-targeting siRNA. At 24 hr post-nucleofection, an intracellular anti-HA staining was analyzed by flow cytometry. Fold change by US10 siRNA is depicted in (E) Short exposure of HLA-I HCs from the immunoprecipitation experiment shown in Figure 6B (Figure 6—figure supplement 1—source data 2).
-
Figure 6—figure supplement 2—source data 1
Uncropped blots from Figure 6—figure supplement 2B.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig6-figsupp2-data1-v1.zip
-
Figure 6—figure supplement 2—source data 2
Uncropped version of Figure 6—figure supplement 2E.
- https://cdn.elifesciences.org/articles/85560/elife-85560-fig6-figsupp2-data2-v1.zip
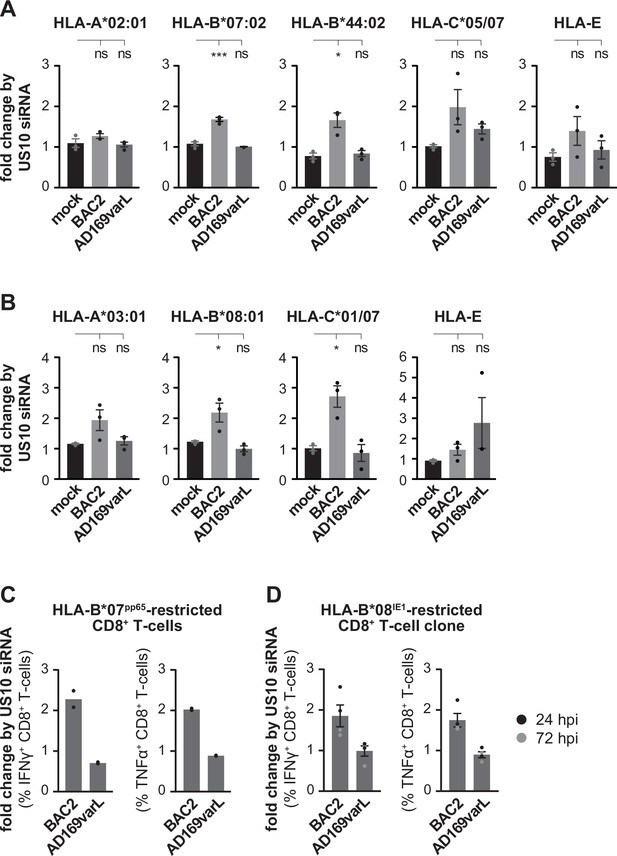
US10 siRNA treatment of human cytomegalovirus (HCMV)-infected cells has little effect on HLA-A, but induce HLA-B1027 antigen presentation.
(A, B) MRC-5 (A) or HF99/7 (B) fibroblasts were nucleofected with US10-specific or non-targeting siRNA 24 hr prior to mock treatment or infection at an MOI of 5 with HCMV ΔUS2-6 mutant BAC2 or with AD169varL. At 48 h p.i., HLA-I surface expression was measured by flow cytometry using antibodies as indicated. Fold change by US10 was calculated as the ratio of the median fluorescence intensity (MFI) of cells treated with US10 siRNA compared to NT-treated cells. Dots represent individual values and bars mean values ± SEM from three independent experiments (biological replicates). Significance was calculated using one-way paired ANOVA followed by Dunnett’s multiple comparison test. (C, D) HFFα (C) or HFF99/7 (D) fibroblasts were treated and infected as in (A, B). At 24 h p.i., the fibroblasts were co-cultured for 5 hr with HLA-B*07:02pp65-specific polyclonal CD8+ T-cells gained from PBMCs (peripheral blood mononuclear cells) (C) or with an HLA-B*08IE1-specific CD8+ T-cell clone at an E/T ratio of 3:1 (C) and 5:1 (D), respectively. Activation of CD8+ T-cells was determined by intracellular IFNγ and TNFα stain. The percentage of IFNγ- or TNFα-expressing CD8+ T-cells was measured and the fold change by US10 siRNA was calculated. Dots represent individual values from two (C) and four (D) independent experiments (biological replicates). Co-culturing for (D) took place 24 (black dots) or 72 hr p.i. (gray dots). Representative dot plots are shown in Figure 7—figure supplement 2A and B.
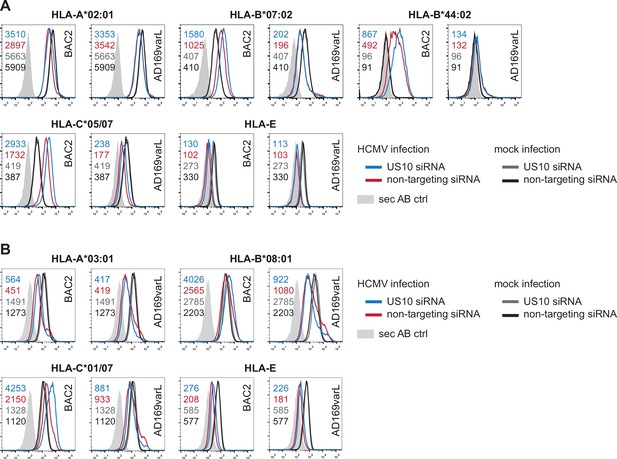
Human leucocyte antigen class I (HLA-I) analysis of US10 siRNA-treated human cytomegalovirus (HCMV)-infected fibroblasts.
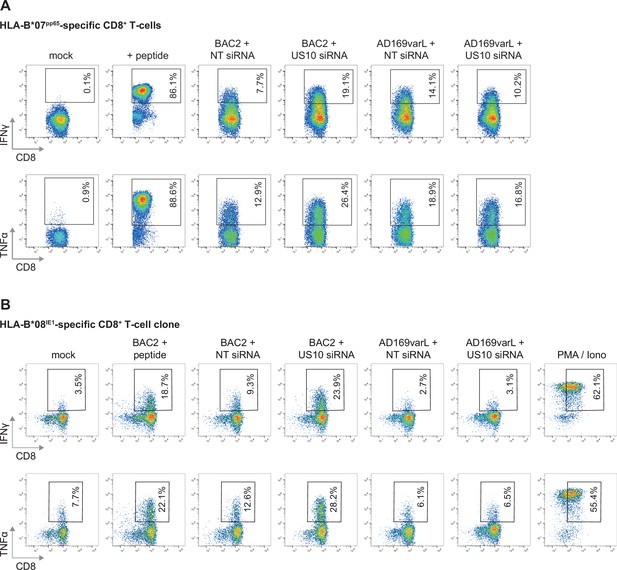
CD8+ T-cell activation after co-culture with US10 siRNA-treated human cytomegalovirus (HCMV)-infected fibroblasts.
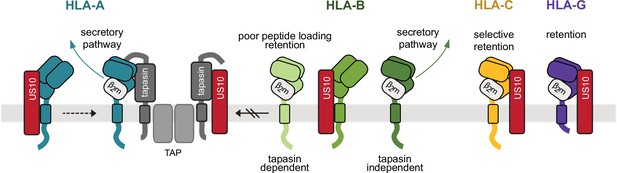
Model of human leucocyte antigen class I (HLA-I) geno- and allotype-dependent targeting by US10.
US10 (red) is able to bind to all HLA-I heavy chains (HCs) early after their synthesis and prior to dimerization with β2m. HLA-A (blue) and HLA-B (green colors) molecules can escape from US10 by dimerization with β2m. In addition, US10 blocks HLA-I recruitment to the peptide loading complex (PLC). This has a pronounced inhibitory effect on tapasin-dependent HLA-B allotypes. HLA-A allotypes can overcome this inhibition. β2m-assembled HLA-G (purple) and some -C (yellow) molecules are strongly retained in the endoplasmic reticulum (ER).
Tables
Primer sequences for molecular cloning.
| HA-HLA-A*24:02 |
|
| HA-HLA-C*04:01 |
|
| HA-HLA-C*05:01 |
|
| HA-HLA-G*01:01 |
|
| HA-H-2Kb |
|
| HA-MICA*004 |
|
| HLA-A2E*01:01 |
|
| ΔCRISPR2-HA-HLA-A*02:01 |
|
| ΔCRISPR2-HA-HLA-B*07:02 |
|
| ΔCRISPR2-HA-HLA-B*44:02 |
|
| ΔCRISPR2-HA-HLA-C*05:01 |
|
| ΔCRISPR2-HA-HLA-C*07:02 |
|
| ΔCRISPR2-HA-HLA-E*01:01 |
|
| ΔCRISPR2-HA-HLA-G*01:01 |
|
| RL8 |
|
| US10 |
|
| US10HA |
|
| US2 |
|
| US2HA |
|
| US3HA |
|
| US9HA |
|
| UL40 |
|
-
*
Primers written in italics are identical.





