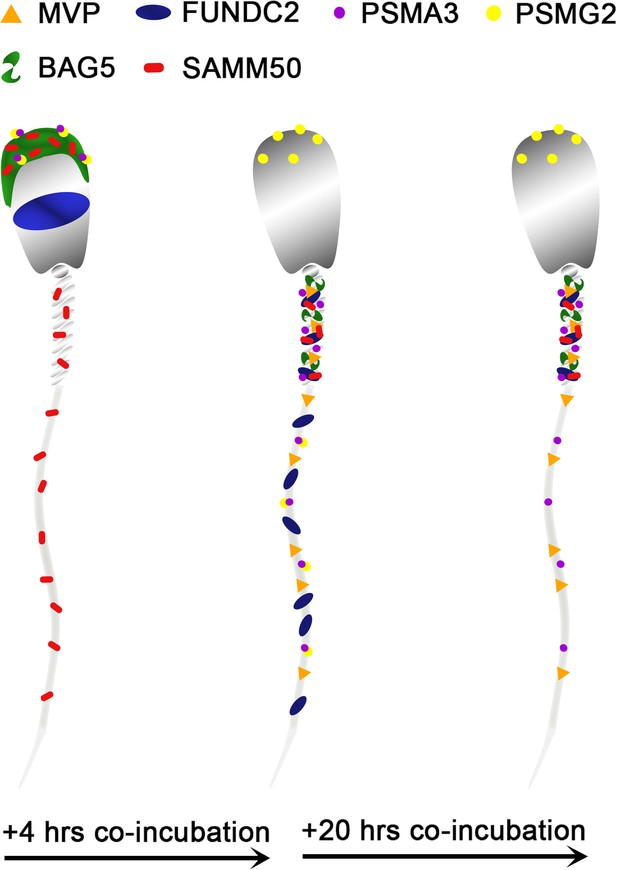
Identification of candidate mitochondrial inheritance determinants using the mammalian cell-free system
Figures
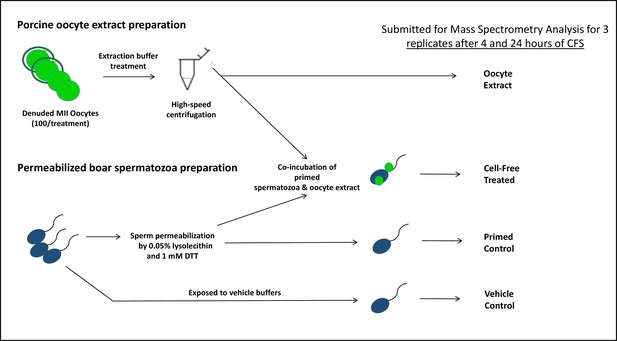
The porcine cell-free system and workflow diagram for the preparation of samples for mass spectrometry analysis.
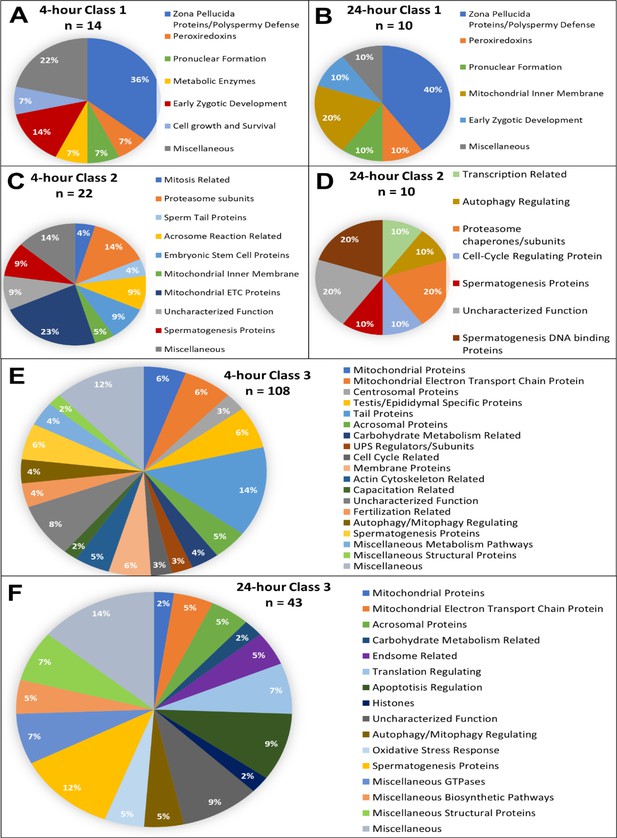
Candidate protein categorization by their functions.
Proteins found in different amounts after cell-free system coincubation at both 4 and 24 hr are categorized by function as found on Uniprot.org and literature via PubMed. Protein characterizations of Class 1 found only in cell-free treated spermatozoa samples after 4 hr of cell-free system co-incubation (A), and 24 hr of cell-free system co-incubation (B) vs primed control spermatozoa (p<0.2). Characterization of Class 2 proteins which underwent an increase in abundance (p<0.1) in cell-free-treated spermatozoa during the 4 hr (C) and 24 hr (D) cell-free system trials vs primed control spermatozoa. Class 3 proteins, which underwent a decrease in abundance (p<0.1) in cell-free-treated spermatozoa after 4 hr (E) and 24 hr (F) of cell-free system co-incubation vs primed control spermatozoa.
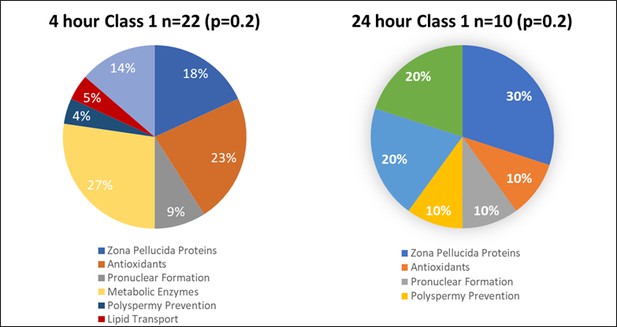
Proteins found in different amounts after Cell-free coincubation at both 4 and 24 hr categorized by function as found on Uniprot.org and literature via Pubmed.com.
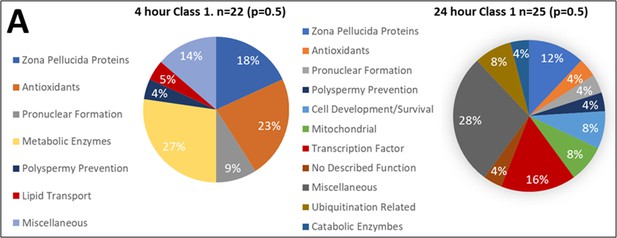
Class 1 protein categories found in higher abundance (p<0.2) after 4 and 24 hr of cell-free coincubation.
Proteins found in different amounts after Cell-free coincubation at both 4 and 24 hr categorized by function as found on Uniprot.org and literature via Pubmed.com.
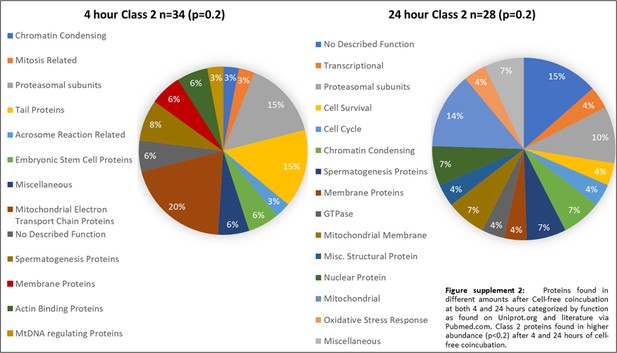
Class 2 proteins found in higher abundance (p<0.2) after 4 and 24 hr of cell-free coincubation.
Proteins found in different amounts after Cell-free coincubation at both 4 and 24 hr categorized by function as found on Uniprot.org and literature via Pubmed.com.
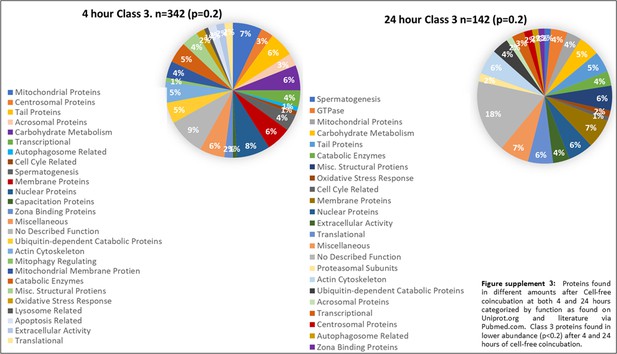
Class 3 proteins found in lower abundance (p<0.2) after 4 and 24 hr of cell-free coincubation.
Proteins found in different amounts after Cell-free coincubation at both 4 and 24 hr categorized by function as found on Uniprot.org and literature via Pubmed.com.
Class 1 protein categories found in higher abundance (p<0.3) after 4 and 24 hr of cell-free coincubation.
Proteins found in different amounts after Cell-free coincubation at both 4 and 24 hr categorized by function as found on Uniprot.org and literature via Pubmed.com.
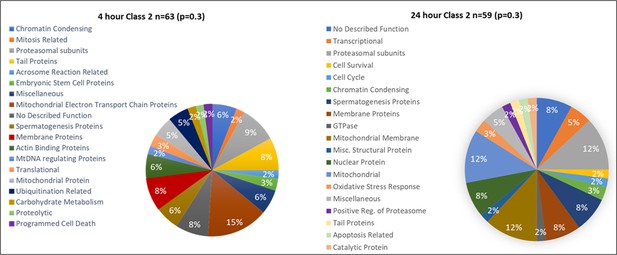
Class 2 proteins found in higher abundance (p<0.3) after 4 and 24 hr of cell-free coincubation.
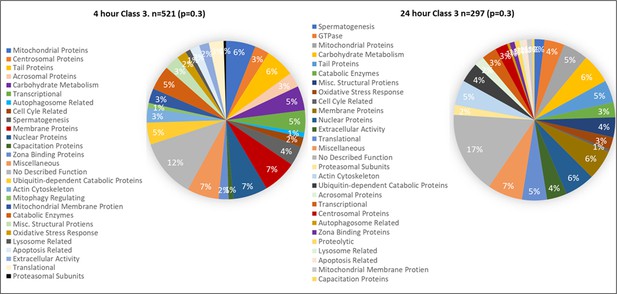
Proteins found in different amounts after Cell-free coincubation at both 4 and 24 hr categorized by function as found on Uniprot.org and literature via Pubmed.com.
Class 3 proteins found in lower abundance (p<0.3) after 4 and 24 hr of cell-free coincubation.
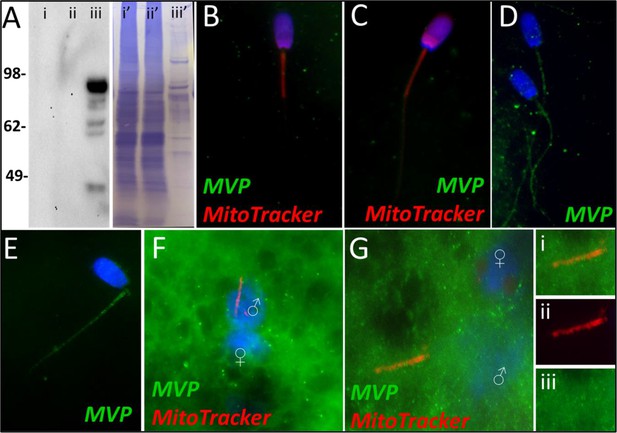
MVP in the porcine cell-free system.
Major vault protein was not detected in ejaculated (Lane i) or capacitated spermatozoa (Lane ii) via Western blot detection, but it was detected in oocyte extract (Lane iii) (A; predicted mass = 99 kDa); (Lanes i’, ii’, iii’) On the right side of panel A, PVDF membrane stained with Coomassie brilliant blue after chemiluminescence detection shows protein loads within each lane. MVP was not detected in ejaculated (B) or primed spermatozoa (C) via immunocytochemistry. After both 4 and 24 hr of cell-free system exposure, MVP (green) was then detected throughout the tail of the treated spermatozoa (D and E). MVP was detected in the cytoplasm of zygotes 15 hr post insemination, but did not appear to associate with the pronuclei or mitochondrial sheath (F). MVP was still detected in the cytoplasm of zygotes 25 hr post insemination, and did appear to have some association with the mitochondrial sheath of the fertilizing spermatozoa (G). A zoomed-in cutout of the mitochondrial sheath can be found in (Gi). The mitochondrial sheath is shown by MitoTracker labeling in the red channel separation panel (Gii). The green/protein labeling channel separation is shown in (Giii).
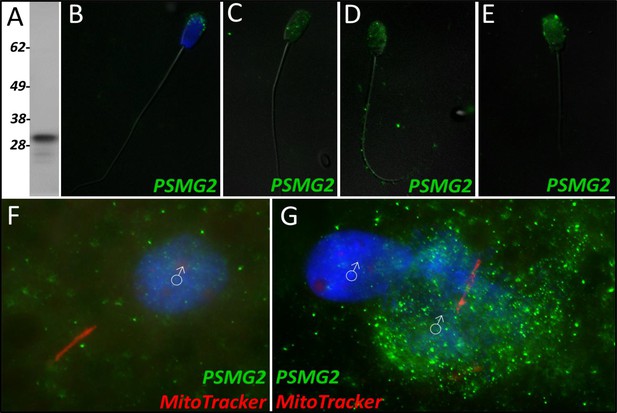
PSMG2 in the porcine cell-free system.
Proteasomal assembly chaperone 2 was detected in ejaculated spermatozoa using Western blotting (A; predicted mass = 29 kDa), and immunocytochemistry (green) (B), where it was found to localize to the acrosome. In primed spermatozoa, PSMG2 was spread throughout the entire head (C). After 4 hr of cell-free system exposure, this same localization pattern was detected in the sperm head, but PSMG2 was also localized to the principal piece of the tail (D). After 24 hr of cell-free system exposure, PSMG2 was only detected in the head of the treated spermatozoa (E). PSMG2 was detected around the newly forming paternal pronuclei in zygotes 15 hr post insemination (F). PSMG2 is detected robustly localizing to the paternal pronuclei and the mitochondrial sheath of the fertilizing spermatozoa in this polyspermic zygote, 25 hr post insemination (G). Note that the larger, more developed PN on the right has the majority of the PSMG2 labeling.
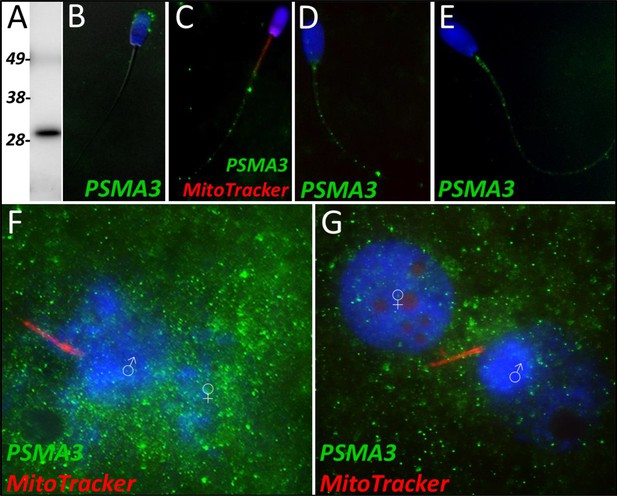
PSMA3 in the porcine cell-free system.
Proteasomal subunit alpha 3 was detected in ejaculated spermatozoa via Western blotting detection (A; predicted mass = 28.4 kDa). PSMA3 (green) was localized to the acrosome of ejaculated spermatozoa by immunocytochemistry (B). After the priming process, PSMA3 was detected in the tail of primed spermatozoa (C). This same tail localization pattern was observed after both 4 and 24 hr of cell-free system exposure (D and E). PSMA3 was detected in zygotes both 15 and 25 hr post insemination (F and G); it was detected surrounding the male and female pronuclei as well as on the mitochondrial sheath of the fertilizing spermatozoa.
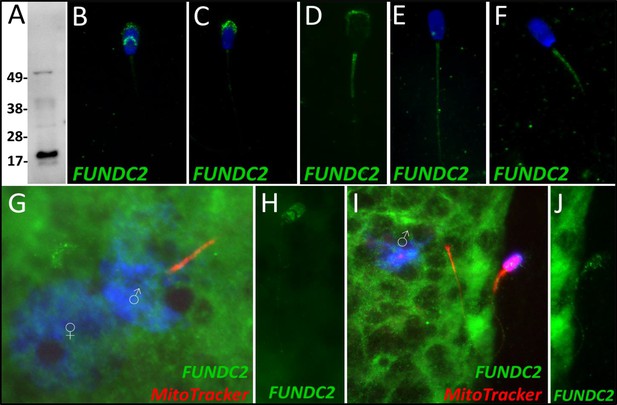
FUNDC2 in the porcine cell-free system.
FUN14 domain-containing protein 2 was detected in ejaculated spermatozoa by Western blotting (A; predicted mass = 20.7 kDa). FUNDC2 was detected in the acrosome and equatorial segment of ejaculated spermatozoa (B) and the acrosome and mitochondrial sheath of capacitated spermatozoa (C). In primed spermatozoa, FUNDC2 was detected in the mitochondrial sheath and the remnants of the acrosome (D). After 4 hr of cell-free system exposure, FUNDC2 was detected throughout the tail of the treated spermatozoa (E), and after 24 hr of cell-free system exposure, FUNDC2 was detected in the mitochondrial sheath of treated spermatozoa (F). FUNDC2 was not detected localizing to the fertilizing sperm components in zygotes 15 hr post insemination (G). In contrast, a zona-bound spermatozoon at that same time point had FUNDC2 localized throughout the head and some tail localization as well (H). FUNDC2 was not detected localizing to the fertilizing sperm components in zygotes 25 hr post insemination (I), but a non-fertilizing spermatozoon bound to the oolemma at this same time point still had some FUNDC2 labeling in its tail and head, as shown by a fluorescent channel separated cutout (J).
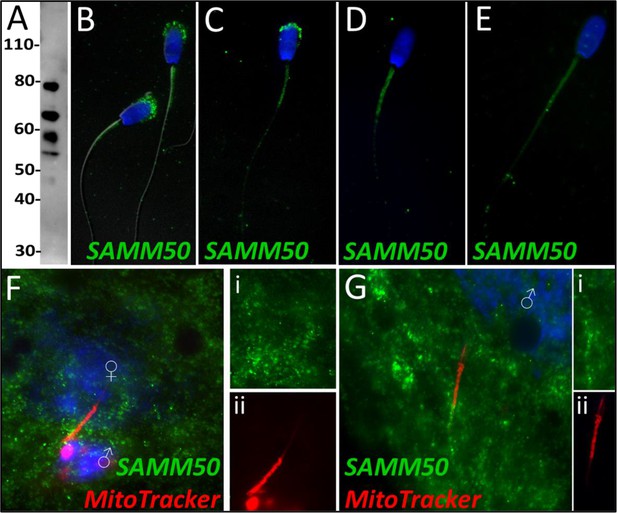
SAMM50 in the porcine cell-free system.
Sorting and assembly machinery component 50 was detected in ejaculated spermatozoa using Western blotting (A; predicted mass = 51 kDa). SAMM50 has predicted post translational modifications, including phosphorylation sites and ubiquitination sites, as predicted using the MuSite Deep prediction software which likely explains the higher bands which are observed. SAMM50 was also detected in ejaculated spermatozoa using immunocytochemistry and found to localize to the acrosome and the mitochondrial sheath of the sperm tail (B). In primed spermatozoa, SAMM50 was detected in the remnants of the acrosome, as well as in the mitochondrial sheath, and the principal piece (C). After 4 hr of cell-free system exposure, SAMM50 was detected predominantly in the mitochondrial sheath with some signal present in the principal piece of the sperm tail as well (D). After 24 hr of cell-free system exposure, SAMM50 was detected predominantly in the mitochondrial sheath, still with residual principal piece labeling as well (E). SAMM50 in zygotes 15 hr post insemination was detected on and near the mitochondrial sheath of the fertilizing spermatozoa (F). Fluorescence channel separation cutouts of the mitochondrial sheath and principal piece are shown in (Fi; green SAMM50) and (Fii; red MitoTracker). SAMM50 was also detected in zygotes 25 hr post insemination on the principal piece of the tail of the fertilizing spermatozoa, just below the mitochondrial sheath (G). Fluorescence channel separation cutouts of the mitochondrial sheath and principal piece remnants are shown in (Gi; green SAMM50) and (Gii; red MitoTracker).
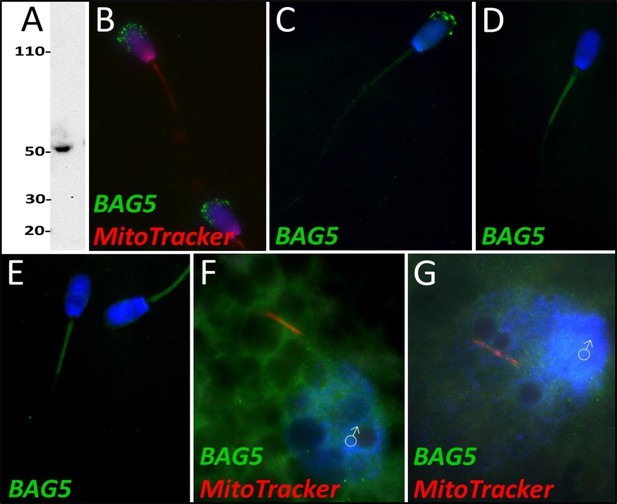
BAG family molecular chaperone regulator 5 was detected in ejaculated spermatozoa via WB (A; predicted mass = 54 kDa) and immunocytochemistry (B), where it was detected to localize in the acrosome.
After priming, BAG5 still remained localized to what remained of the acrosome (C). After 4 and 24 hr of cell-free system exposure, BAG5 was detected in the mitochondrial sheath of the treated spermatozoa (D and E). BAG5 was detected in the cytoplasm in zygotes 15 hr post structures (F) and 25 hr post fertilization, with no obvious association with the fertilizing sperm structures or their remnants (G).
Additional files
-
Supplementary file 1
Raw data captured from mass spectrometry, referenced against the Sus scrofa UniProt Knowledge base.
- https://cdn.elifesciences.org/articles/85596/elife-85596-supp1-v1.xlsx
-
Supplementary file 2
Proteomic identification of mitophagy and sperm remodeling cofactors in the porcine cell-free system after 4 hours of co-incubation.
- https://cdn.elifesciences.org/articles/85596/elife-85596-supp2-v1.docx
-
Supplementary file 3
Proteomic identification of mitophagy and sperm remodeling cofactors in porcine cell-free system after 24 hours of co-incubation.
- https://cdn.elifesciences.org/articles/85596/elife-85596-supp3-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85596/elife-85596-mdarchecklist1-v1.docx