Astrocyte and L-lactate in the anterior cingulate cortex modulate schema memory and neuronal mitochondrial biogenesis
Figures
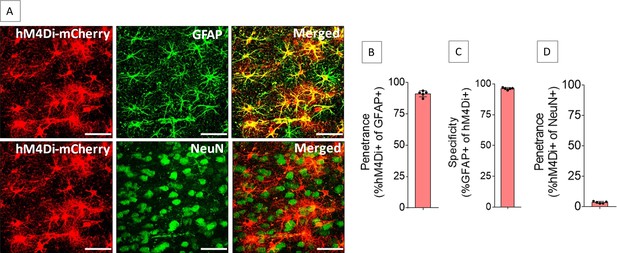
Expression of hM4Di in anterior cingulate cortex (ACC) astrocytes.
Injection of AAV8-GFAP-hM4Di-mCherry into the ACC resulted in expression of hM4Di (A) in 91.1 ± 2.4% of GFAP-positive cells (B) with 96.3 ± 0.9% specificity (C), whereas 3.6 ± 0.8% of NeuN-positive cells expressed hM4Di (D). n=5 rats (3 sections/rat). Scale bars: 50 μm.
-
Figure 1—source data 1
Zip file containing data for Figure 1B–D in GraphPad Prism file format.
- https://cdn.elifesciences.org/articles/85751/elife-85751-fig1-data1-v1.zip
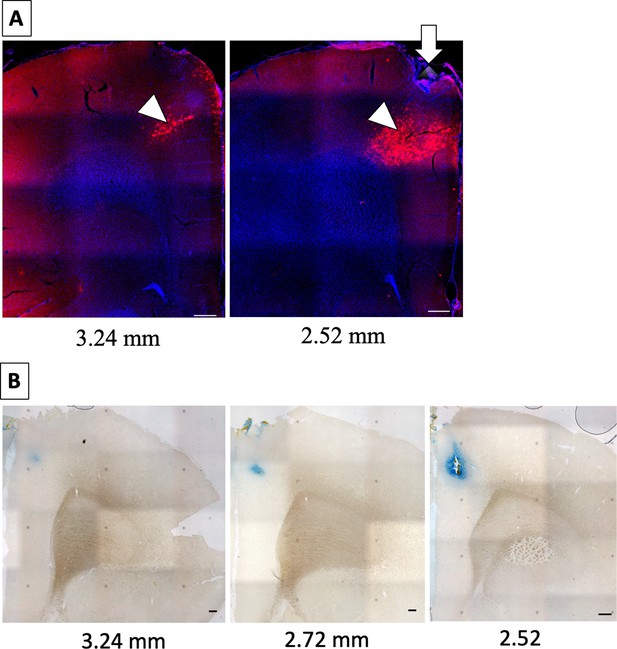
Microinjection and cannulation in the anterior cingulate cortex (ACC).
(A) Representative sections 3.24 mm and 2.52 mm anterior to bregma showing hM4Di-mCherry expression (arrowheads) in the ACC. The arrow shows the site of microinjection and cannula placement into the ACC. Scale bars: 200 µm. (B) Representative sections 3.24 mm, 2.72 mm, and 2.52 mm anterior to bregma under light microscopy. The rat was injected with 1 μl of Chicago sky blue (1%) into the ACC and sacrificed 1 hr following the injection. Scale bars: 200 µm.
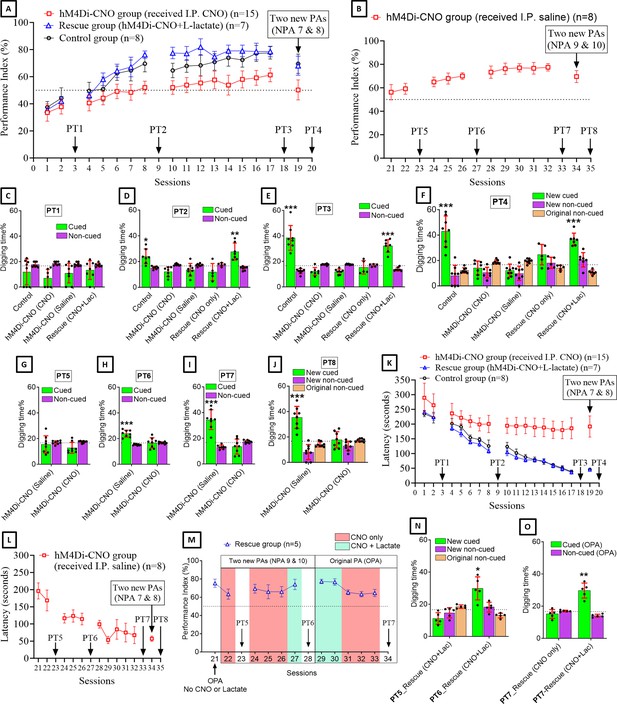
Astrocytic Gi pathway activation in the anterior cingulate cortex (ACC) impairs paired associate (PA) learning, schema consolidation, memory retrieval, and assimilation of new PAs (NPAs) into existing schema whereas L-lactate can rescue these impairments.
(A) Performance index (PI) (mean ± SD) during the acquisition of the six original PAs (OPAs) (S1–2, 4–8, 10–17) and NPAs (S19) of the control (n=8), hM4Di-CNO (n=15), and rescue (hM4Di-CNO+L-lactate) (n=7) groups. From S6 onward, hM4Di-CNO group consistently showed lower PI compared to control. However, concurrent L-lactate administration into the ACC (rescue group) can rescue this impairment. (B) PI (mean ± SD) of hM4Di-CNO group (n=8) from S21 onward showing gradual increase in PI when CNO was withdrawn. (C, D, and E) Non-rewarded PTs (PT1, PT2, and PT3 conducted on S3, S9, and S18, respectively) to test memory retrieval of OPAs for the control, hM4Di-CNO, and rescue groups. The percentage of digging time at the cued location relative to that at the non-cued locations are shown (mean ± SD). In both PT2 and PT3, the control group spent significantly more time digging the cued sand well above the chance level, indicating that the rats learned OPAs and could retrieve it. Contrasting to this, hM4Di-CNO group did not spend more time digging the cued sand well above the chance level irrespective of CNO administration before the PTs. The rescue group showed results similar to the hM4Di-CNO group if CNO is given without L-lactate. On the other hand, they showed results similar to the control group if L-lactate is concurrently given with CNO, indicating that this group learned OPAs and could retrieve it. *p<0.05, **p<0.01, ***p<0.001, one-sample t-test comparing the proportion of digging time at the cued sand well with the chance level of 16.67%. Non-rewarded PT4 (S20) which was conducted after replacing two OPAs with two NPAs (NPAs 7 and 8) in S19 for the control, hM4Di-CNO, and rescue groups. Results show that the control group spent significantly more time digging the new cued sand well above the chance level indicating that the rats learned the NPAs from S19 and could retrieve it in this PT. Contrasting to this, hM4Di-CNO group did not spend more time digging the new cued sand well above the chance level irrespective of CNO administration before the PT. The rescue group showed results similar to the hM4Di-CNO group if CNO is given without L-lactate. On the other hand, they showed results similar to the control group if L-lactate is concurrently given with CNO indicating that this group learned NPAs from S19 and could retrieve it. ***p<0.001, one-sample t-test comparing the proportion of digging time at the new cued sand well with the chance level of 16.67%. (G, H, and I) Non-rewarded PTs (PT5, PT6, and PT7 conducted on S23, S27, and S33, respectively) to test memory retrieval of OPAs for the hM4Di-CNO group. In both PT6 and PT7, the rats spent significantly more time digging the cued sand well above the chance level if the tests are done without CNO, indicating that the rats learned the OPAs and could retrieve it. However, CNO prevented memory retrieval during these PTs. ***p<0.001, one-sample t-test comparing the proportion of digging time at the cued sand well with the chance level of 16.67%. Non-rewarded PT4 (S35) which was conducted after replacing two OPAs with two NPAs (NPAs 9 and 10) in S34 for the hM4Di-CNO group. Results show that the rats spent significantly more time digging the new cued sand well above the chance level if CNO was not given before the PT, indicating that the rats learned the NPAs from S34 and could retrieve it in this PT. However, if CNO is given before the PT, the retrieval is impaired. ***p<0.001, one-sample t-test comparing the proportion of digging time at the new cued sand well with the chance level of 16.67%. (K and L) Latency (in seconds) before commencing digging at the correct well. Data shown as mean ± SD. (M, N, and O) Continuation study (S21–34) with the rescue group (n=5). The PI (mean ± SD) is shown in (M). PT5 and PT6 (conducted at S23 and S28, respectively) are shown in (N). PT7, which was conducted twice, is shown in (O). In S21, PI was 75.3 ± 4.5% for the six OPAs without CNO or L-lactate. For S22–28, two OPAs were replaced with two NPAs (NPAs 9 and 10). In S22, which was conducted with CNO only, PI dropped to 63.3 ± 5.6%. PT5 (N) confirms that the rats did not learn the NPAs 9 and 10 from S21. In S24–26, which were conducted with CNO only, PI remained similarly low (69.3 ± 4.9%, 66 ± 7.7%, and 66 ± 5.7%, respectively), indicating that the rats were not learning the NPAs 9 and 10 despite multiple sessions. In S27, which was conducted with CNO+L-lactate, PI raised to 74 ± 5.7%, suggesting that they have learned the NPAs in this session. This was confirmed by PT6 (N) which showed that they spent significantly more time in digging the new cued sand well above the chance level. In S29–34, the six OPAs were restored. Studies in these sessions showed that PI drops from ∼77% to ∼64% even for the OPAs if L-lactate is not given concurrently with CNO. Furthermore, PT7 (S34) (O) shows that CNO administration before PT impairs memory retrieval of existing associative schema which can be rescued by administering L-lactate concurrently. *p<0.05, **p<0.01, one-sample t-test comparing the proportion of digging time at the cued sand well with the chance level of 16.67%.
-
Figure 2—source data 1
Zip file containing data for Figure 2A–O, Figure 2—figure supplement 2A–D, and Figure 2—figure supplement 3E–G in GraphPad Prism file format.
- https://cdn.elifesciences.org/articles/85751/elife-85751-fig2-data1-v1.zip
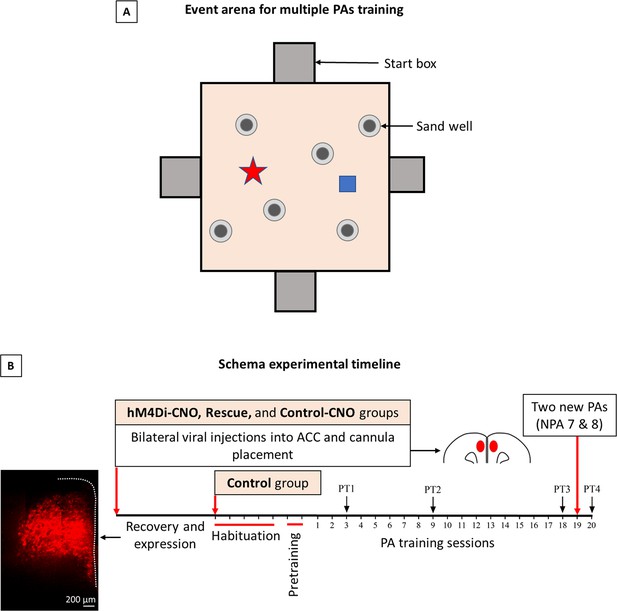
Schema experimental design.
(A) Event arena for multiple flavor-place paired associate training. (B) Timeline for schema experiments.

Clozapine-N-oxide (CNO) application itself has no effect on paired associate (PA) learning and memory retrieval.
(A) Performance index (PI) (mean ± SD) during the acquisition of the six original PAs (OPAs) (S1–2, 4–8, 10–17) and new PAs (NPAs) (S19) of the control (n=6) and control-CNO (n=4) groups. The control group data is the same data as shown in Figure 2A. (B) Non-rewarded PTs (PT1, PT2, and PT3 conducted on S3, S9, and S18, respectively) to test memory retrieval of OPAs for the control-CNO group. (C) Non-rewarded PT4 (S20) which was conducted after replacing two OPAs with two NPAs (NPAs 7 and 8) in S19 for the control-CNO group. (D) Latency (in seconds) before commencing digging at the correct well for the control and control-CNO groups. Data shown as mean ± SD. The control group data is the same data as shown in Figure 2K.

Results of the open field test (OFT).
(A) Timeline for the experiments. OFT was conducted for all groups (hM4Di-saline, hM4Di-CNO, and hM4Di-CNO+L-lactate; n=8 in each group) after two paired associate (PA) training sessions. Additionally, OFT was conducted after S8 and S17 of PA training sessions for the hM4Di-CNO group. (B–D) Representative track plots (300 s) of hM4Di-saline, hM4Di-CNO, and hM4Di-CNO+L-lactate groups. The inner square within the plots marks the central zone. (E–G) Figures showing the distance traveled (in meters), time spent in the central zone (in seconds), and number of entries into the central zone in 300 s, respectively.
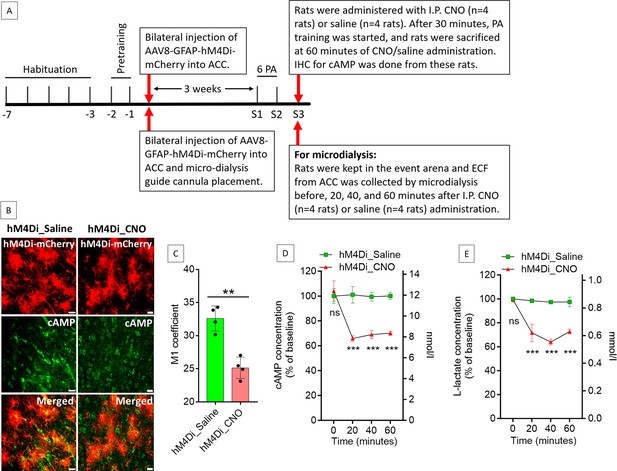
Effect of anterior cingulate cortex (ACC) astrocytic Gi activation on cyclic adenosine monophosphate (cAMP) and L-lactate levels in the ACC.
(A) Experimental design to investigate the effect of Gi activation of ACC astrocytes on cAMP and L-lactate levels. (B and C) CNO decreases cAMP in the hM4Di-expressed cells. (B) Confocal micrograph of ACC 60 min after intraperitoneal administration of saline or CNO in hM4Di-expressed rats. Scale bars: 20 μm. (C) Colocalization analysis showing decreased Mander’s coefficient M1 (ratio of cAMP intensity colocalized with hM4Di-mCherry to total cAMP intensity) in CNO administered rats (n=4 rats in each group; 3 sections/rat). **p=0.001, unpaired Student’s t-test, t=6.01, df = 6. (D and E) Microdialysis measurement of cAMP (D) and L-lactate (E) levels in the extracellular fluid (ECF) of ACC before, 20 min, 40 min, and 60 min after intraperitoneal saline or CNO administration in hM4Di-expressed rats (n=4 rats in each group). ns = not significant, ***p<0.001, unpaired Student’s t-test.
-
Figure 3—source data 1
Zip file containing data for Figure 3C–E in GraphPad Prism file format.
- https://cdn.elifesciences.org/articles/85751/elife-85751-fig3-data1-v1.zip
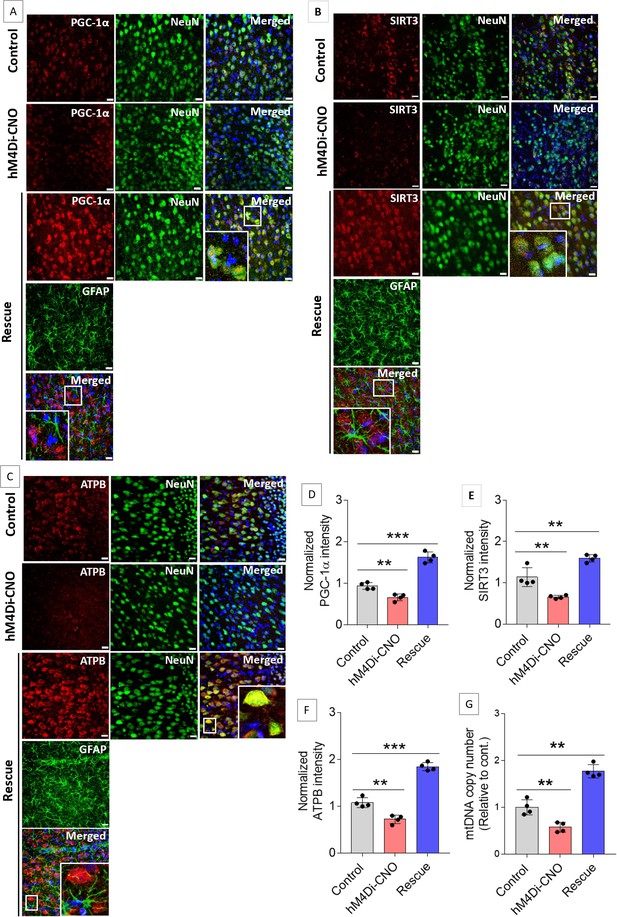
Gi activation in anterior cingulate cortex (ACC) astrocytes reduces neuronal mitochondrial biogenesis whereas concurrent exogenous L-lactate administration rescues the impairment.
(A–C) Representative confocal micrograph of PGC-1α (A)/SIRT3 (B)/ATPB (C) co-labeled with NeuN, glial fibrillary acidic protein (GFAP), and DAPI (4′,6-diamidino-2-phenylindole) in the ACC of the control, hM4Di-CNO, and rescue groups from schema experiments. Astrocytic Gi pathway activation (hM4Di-CNO group) in the ACC resulted in decreased PGC-1α/SIRT3/ATPB expression in the ACC, whereas concurrent exogenous L-lactate (rescue group) administration resulted in increased PGC-1α/SIRT3/ATPB expression. Scale bars: 20 µm. (D–F) Fluorescence intensity of PGC-1α (D)/SIRT3 (E)/ATPB (F) stained sections in the ACC of hM4Di-CNO and rescue groups were assessed and normalized to the control group of rats. Data shown as mean ± SD (n=4 rats per group, 3 sections/rat). **p<0.01, ***p<0.001, unpaired Student’s t-test. (G) mtDNA copy number abundance in the ACC of control, hM4Di-CNO, and rescue groups relative to nDNA. Relative mtDNA copy number was significantly decreased in the hM4Di-CNO group, whereas it was increased in the rescue group compared to control. Data shown as mean ± SD (n=4 rats per group). **p<0.01, unpaired Student’s t-test.
-
Figure 4—source data 1
Zip file containing data for Figure 4D–G in GraphPad Prism file format.
- https://cdn.elifesciences.org/articles/85751/elife-85751-fig4-data1-v1.zip
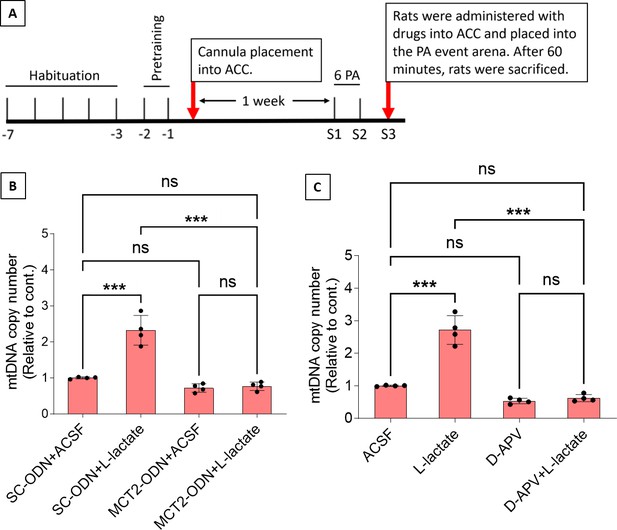
Mitochondrial biogenesis by L-lactate is dependent on monocarboxylate transporter 2 (MCT2) and NMDA receptor (NMDAR).
(A) Experimental design to investigate whether MCT2 and NMDAR activity are required for L-lactate-induced mitochondrial biogenesis. (B and C) mtDNA copy number abundance in the anterior cingulate cortex (ACC) of different rat groups relative to nDNA. Data shown as mean ± SD (n=4 rats in each group). ***p<0.001, ANOVA followed by Tukey’s multiple comparisons test.
-
Figure 5—source data 1
Zip file containing data for Figure 5B–C and Figure 5—figure supplement 1B in GraphPad Prism file format.
- https://cdn.elifesciences.org/articles/85751/elife-85751-fig5-data1-v1.zip

Western blot of monocarboxylate transporter 2 (MCT2).
(A) Western blot showing decreased expression of MCT2 in the anterior cingulate cortex (ACC) 12 hr after injection of MCT2 antisense oligodeoxynucleotide. (B) Intensity of MCT2 was quantified and normalized with β-actin. Data shown as mean ± SD (n = 3 rats per group). **p < 0.01, unpaired Student’s t-test.
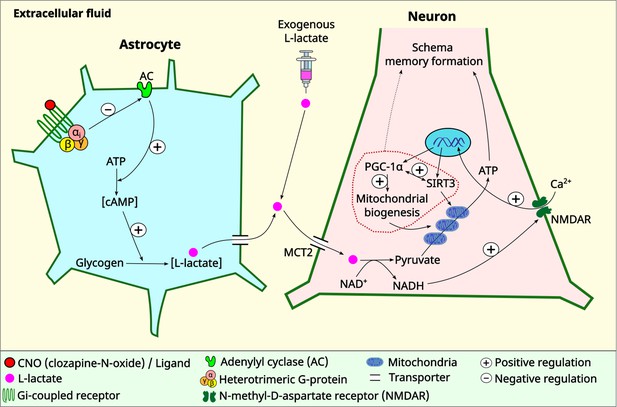
Schematic diagram showing astrocytic Gi signaling and L-lactate modulating schema memory and mitochondrial biogenesis.
L-lactate in the anterior cingulate cortex (ACC) is required for schema memory formation and neuronal mitochondrial biogenesis. Astrocytic Gi activation results in decreased L-lactate in the ACC with consequent impairments in schema memory and neuronal mitochondrial biogenesis which could be rescued by exogenous L-lactate administration directly into the ACC. Further research is needed to establish the mechanism and the extent of the contribution of mitochondrial biogenesis in schema memory formation (dotted arrow). MCT2: monocarboxylate transporter 2.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Rattus norvegicus, male) | Adult Sprague- Dawley rats (250–300 g) | Laboratory Animal Services Centre, Chinese University of Hong Kong, SAR, China | ||
| Transfected construct (Rattus norvegicus) | AAV8-GFAP-hM4Di-mCherry | Shanghai Taitool Bioscience Co. Ltd | ||
| Transfected construct (Rattus norvegicus) | AAV8-GFAP-mCherry | Shanghai Taitool Bioscience Co. Ltd | ||
| Sequence-based reagent | Rat MCT2 antisense oligodeoxynucleotide (ODN), 200 nmol (HPLC purified) | Integrated DNA Technologies (IDT) | Cat #: 107968967 | |
| Sequence-based reagent | Rat Relative scrambled ODN, 200 nmol (HPLC purified) | Integrated DNA Technologies (IDT) | Cat #: 107969138 | |
| Sequence-based reagent | Rat D-loop Forward and Reverse Primer | Integrated DNA Technologies (IDT) | Cat #: 107056074 and Cat #: 107056075 | Used to measure mtDNA by real-time PCR |
| Sequence-based reagent | Rat β-actin Forward and Reverse Primer | Integrated DNA Technologies (IDT) | Cat #: 107056076 and Cat #: 107056077 | Used to measure nDNA by real-time PCR |
| Antibody | Anti-GFAP (Mouse Monoclonal) | Abcam | Cat #: ab4648 | 1:500 (IHC) |
| Antibody | Anti-NeuN (Rabbit Polyclonal) | Merck Millipore | Cat #: AB978 | 1:500 (IHC) |
| Antibody | Anti-mCherry (Chicken Polyclonal) | Abcam | Cat #: ab205402 | 1:1000 (IHC) |
| Antibody | Anti-cAMP (Rabbit Monoclonal) | Abcam | Cat #: ab134901 | 1:500 (IHC) |
| Antibody | Anti-SIRT3 (Rabbit Polyclonal) | Sigma-Aldrich | Cat #: SAB5700222 | 1: 250 (IHC) |
| Antibody | Anti-PGC-1α (Rabbit Polyclonal) | Abcam | Cat #: ab191838 | 1: 500 (IHC) |
| Antibody | Anti-ATPB (Mouse Monoclonal) | Abcam | Cat #: ab14730 | 1: 500 (IHC) |
| Antibody | Anti-MCT2 (Rabbit Polyclonal) | Merck Millipore | Cat #: AB3542 | 1: 500 (WB) |
| Antibody | Anti-β-actin (Mouse Monoclonal) | Immunoway | Cat #: YM3028 | 1: 5000 (WB) |
| Antibody | Alexa Flour 488 (Goat Anti-Mouse Polyclonal) | Thermo Fisher Scientific | Cat #: A11001 | 1:300 (IHC) |
| Antibody | Alexa Flour 594 (Goat Anti-Mouse Polyclonal) | Thermo Fisher Scientific | Cat #: A11032 | 1:300 (IHC) |
| Antibody | Alexa Flour 488 (Goat Anti-Rabbit Polyclonal) | Thermo Fisher Scientific | Cat #: A11034 | 1:300 (IHC) |
| Antibody | Alexa Flour 594 (Goat Anti-Mouse Polyclonal) | Thermo Fisher Scientific | Cat #: A11037 | 1:300 (IHC) |
| Antibody | Goat Anti-Rabbit Secondary Antibody, HRP, Polyclonal | Invitrogen | Cat #: 31460 | 1: 5000 (WB) |
| Antibody | Goat Anti-Mouse Secondary Antibody, HRP, Polyclonal | Invitrogen | Cat #: 62–6520 | 1: 5000 (WB) |
| Commercial assay or kit | Lactate Fluorescence Assay kit | Abcam | Cat #: ab65331 | |
| Commercial assay or kit | cAMP complete ELISA kit | Abcam | Cat #: ab133051 | |
| Commercial assay or kit | QIAamp DNA Mini Kits | QIAGEN | Cat #: 1725270 | |
| Commercial assay or kit | SsoAdvanced Universal SYBR Green Supermix | Bio-Rad | Cat #: 1725270 | |
| Commercial assay or kit | RIPA Buffer | Sigma-Aldrich | Cat #: 20-188 | |
| Commercial assay or kit | Phosphatase and protease inhibitor cocktail | Sigma-Aldrich | ||
| Commercial assay or kit | Bradford assay | Bio-Rad | Cat #: 5000205 | |
| Commercial assay or kit | Western Bright ECL HRP substrate | Advansta | Cat #: K12045-D20 | |
| Chemical compound, drug | Clozapine-N-oxide (CNO) dihydrochloride | Hello Bio | Cat #: HB6149 | |
| Chemical compound, drug | NaCl | Sigma-Aldrich | Cat #: S3014-1kg | |
| Chemical compound, drug | L-lactate | Sigma-Aldrich | Cat #: L-7022 | |
| Chemical compound, drug | D-(-)-2-Amino-5-Phosphonopentanoic acid (D-APV) | Sigma-Aldrich | Cat #: A8054 | |
| Chemical compound, drug | Artificial cerebrospinal fluid (ACSF) | Harvard Apparatus | Cat #: 597316 | |
| Chemical compound, drug | Dorminal 20% | Alfasan International BV | Cat #: 013003 | |
| Chemical compound, drug | Urethane | Sigma-Aldrich | Cat #: U2500-500G | |
| Software, algorithm | FIJI ImageJ | National Institutes of Health, Bethesda, MD, USA | ||
| Software, algorithm | Prism GraphPad | GraphPad Software, San Diego, CA, USA | Version 10 | |
| Software, algorithm | Excel | Microsoft | ||
| Other | Microdialysis guide cannula (CMA 11 elite) and probe (3 mm membrane) | CMA Inc | Used to collect ECF from ACC for L-lactate and cAMP assay | |
| Other | Stainless steel guide cannulae, OD 0.41 mm-27G/C | RWD Life Science | Cat #: 62069 | Used in drug and ODN delivery into ACC |
| Other | Dummy cannulae | RWD Life Science | Cat #: 62169 | Used in drug and ODN delivery into ACC |
| Other | Brain slicer | Braintree Scientific, Braintree, MA, USA | Used to collect ACC from whole brain | |
| Other | Stereotaxic frame | RWD | Used to fix head of rats during surgeries | |
| Other | 33-Gauge metal needle, 10 μl micro-syringe | Hamilton, NV, USA | Used in AAV and drug delivery | |
| Other | Microinjection pump | World Precision Instruments, USA | Used in AAV delivery |
Additional files
-
Supplementary file 1
Comparison of performance index of control vs. hM4Di-CNO group.
- https://cdn.elifesciences.org/articles/85751/elife-85751-supp1-v1.docx
-
Supplementary file 2
Comparison of performance index of control vs. rescue group.
- https://cdn.elifesciences.org/articles/85751/elife-85751-supp2-v1.docx
-
Supplementary file 3
Primer sequences and preparation of 20 μl reaction mixture for real-time PCR.
- https://cdn.elifesciences.org/articles/85751/elife-85751-supp3-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85751/elife-85751-mdarchecklist1-v1.docx





