A rapid phylogeny-based method for accurate community profiling of large-scale metabarcoding datasets
Figures
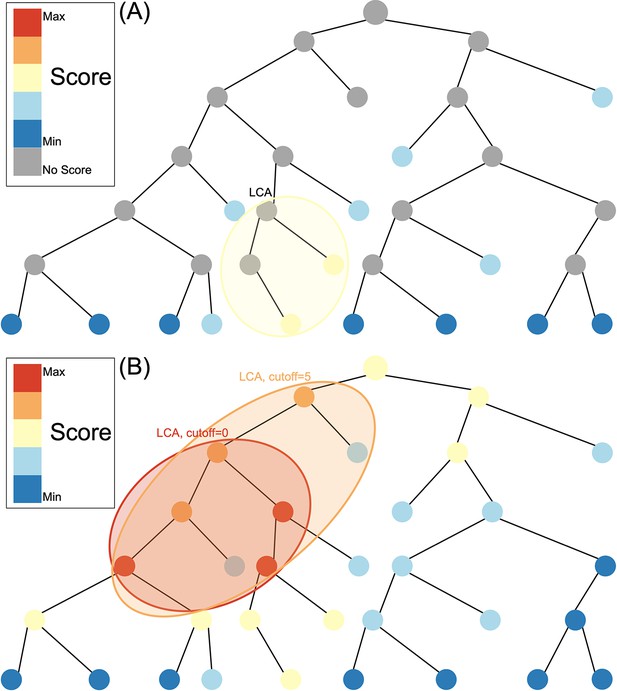
Species assignment in alignment-based methods (A) vs. Tronko (B).
In Tronko, scores are calculated for all nodes in the tree based on the query’s global alignment to the best BWA-MEM hit. The query is assigned to the lowest common ancestor (LCA) of the highest scoring nodes within the cut-off threshold. See Figure 1—figure supplement 1 for more details regarding using multiple trees.
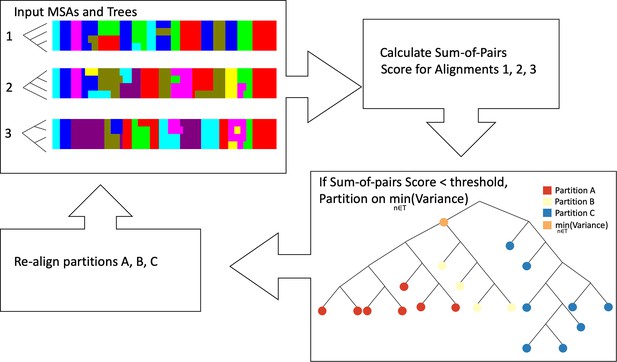
Workflow of iterative partitioning procedure.
First, the multiple sequence alignments (MSAs) and corresponding phylogenetic trees are used as input into the algorithm. Then, the sum-of-pairs scores are calculated for each partition. If the sum-of-pairs score is below a heuristic threshold, the tree is used to partition the sequences into three partitions in the cluster based on the node with the smallest variance. Each of the three partitions is realigned and phylogenetic trees are estimated. The algorithm stops for a given partition when the sum-of-pairs score is greater than the heuristic threshold.
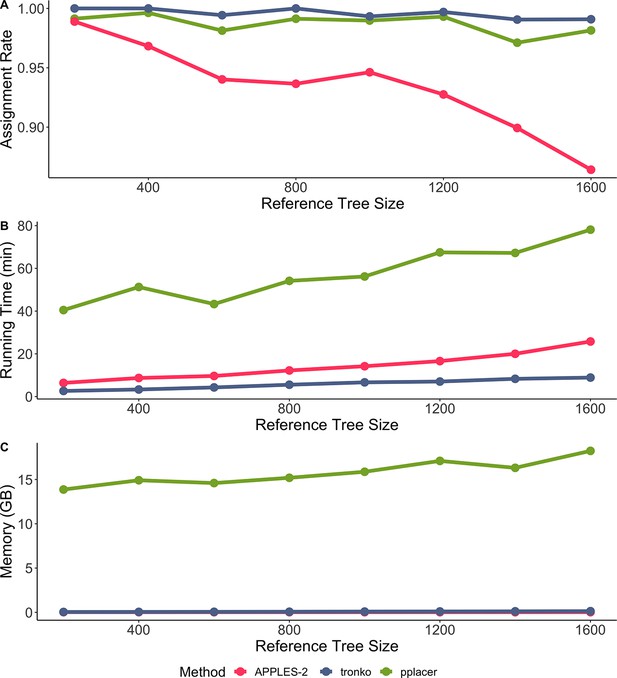
Comparisons of Tronko with pplacer and APPLES-2 using a database of 200, 400, 600, 800, 1000, 1200, 1400, and 1600 reference sequences.
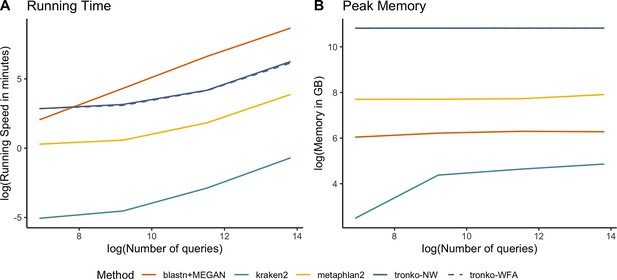
(A) Assignment rate against the number of reference sequences at the species level. (B) Running time against the number of reference sequences. (C) Peak memory in gigabytes against number of references. Both methods had a 100% true positive rate for all sizes of databases. Assignment rate is the number of reads assigned at the species level for each method. Reference sequences were chosen randomly from the cytochrome oxidase 1 (COI) reference database.
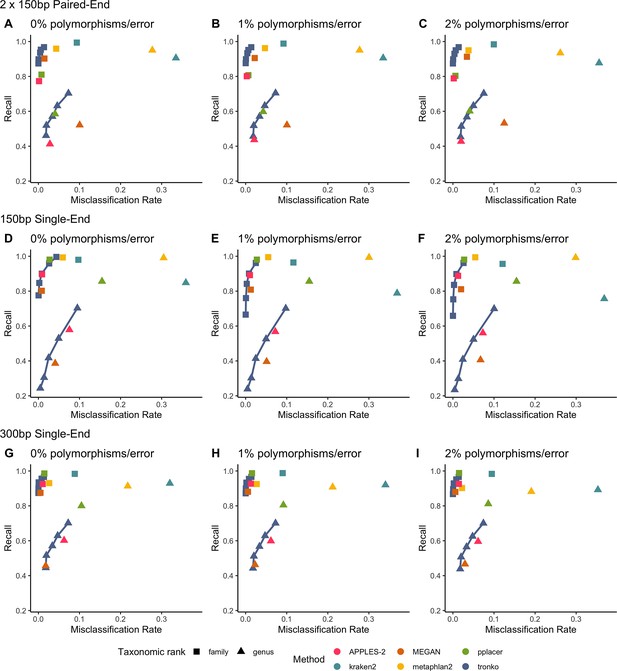
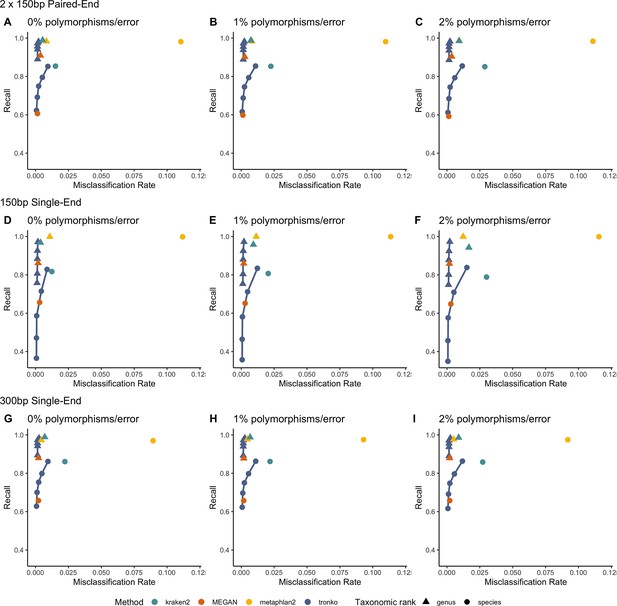
Recall vs. misclassification rates using leave-one-species-out analysis of the order Charadriiformes (cytochrome oxidase 1 [COI] metabarcode) with paired-end 150 bp × 2 reads with 0% (A), 1% (B), and 2% (C) error/polymorphism, single-end 150 bp reads with 0% (D), 1% (E), and 2% (F) error/polymorphism, and single-end 300 bp reads with 0% (G), 1% (H), and 2% (I) error/polymorphism using kraken2, metaphlan2, MEGAN, pplacer, and APPLES-2, and Tronko with cut-offs of 0, 5, 10, 15, and 20 using the Needleman–Wunsch alignment (solid line).
See Figure 2—figure supplement 2 for results using different combinations of aligners and tree estimation methods.
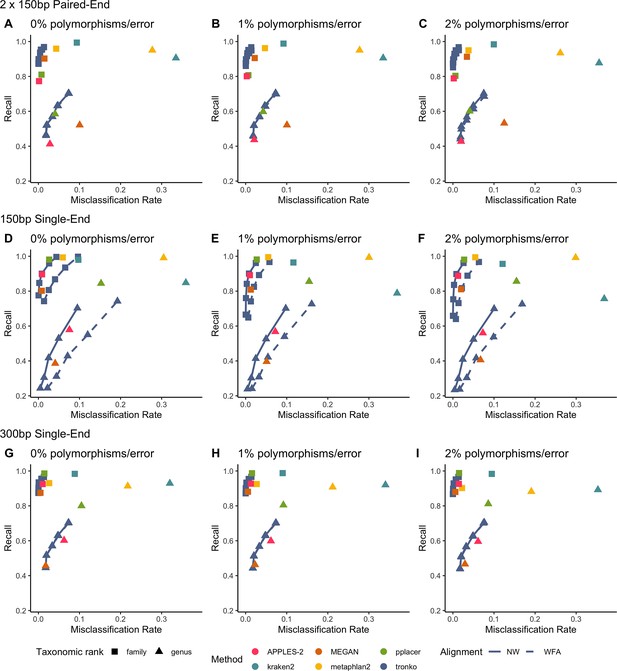
Recall vs. misclassification rates using leave-one-species-out analysis for the order Charadriiformes (cytochrome oxidase 1 [COI] metabarcode) with paired-end 150 bp × 2 reads with 0% (A), 1% (B), and 2% (C) error/polymorphism, single-end 150 bp reads with 0% (D), 1% (E), and 2% (F) error/polymorphism, and single-end 300 bp reads with 0% (G), 1% (H), and 2% (I) error/polymorphism using kraken2, metaphlan2, MEGAN, pplacer, APPLES-2, and Tronko with cut-offs of 0, 5, 10, 15, and 20 using the Needleman–Wunsch alignment (solid line) and wavefront alignment (dashed line).
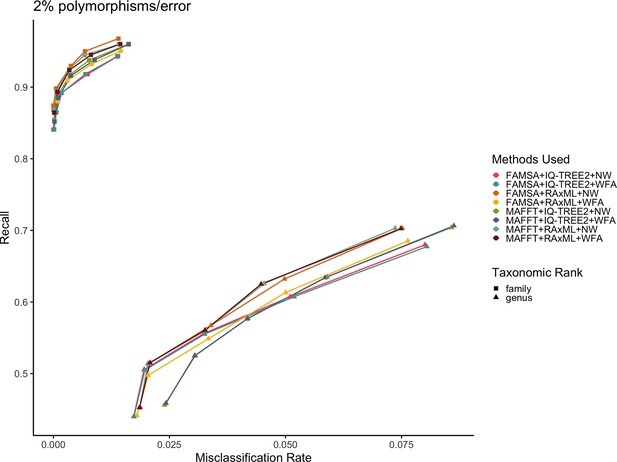
Recall vs. misclassification rates using leave-one-species-out analysis for the order Charadriiformes (cytochrome oxidase 1 [COI] metabarcode) with paired-end 150 bp × 2 reads with 2% error/polymorphism using Tronko with cut-offs of 0, 5, 10, 15, and 20 and different combinations of tree estimation methods and aligners.
For tree estimation, we used RAxML and IQ-TREE2. For multiple sequence aligners, we used FAMSA and MAFFT. For global alignment methods, we used Needleman–Wunsch (NW) and wavefront alignment (WFA). This is the same dataset as used in Figure 2. Colors represent different combinations of methods.
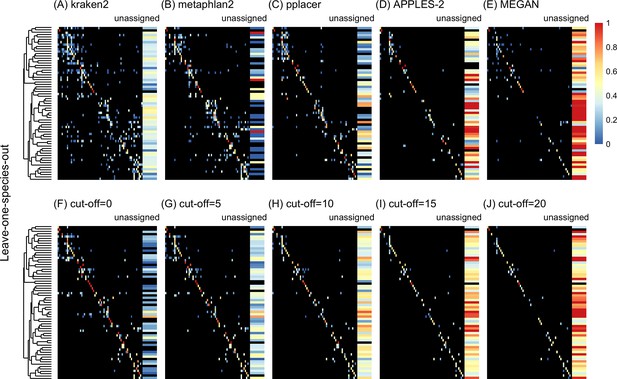
Confusion matrices at the genus level of the order Charadriiformes (cytochrome oxidase 1 [COI] metabarcode) using the leave-one-species-out analysis with paired-end 150 bp × 2 reads with 2% error/polymorphism using kraken2 (A), metaphlan2 (B), pplacer (C), APPLES-2 (D), MEGAN (E), and Tronko using the Needleman–Wunsch alignment (NW) for cut-offs 0 (F), 5 (G), 10 (H), 15 (I), and (J) 20.
Unassigned column contains both unassigned queries and queries assigned to a lower taxonomic level. Phylogenetic tree represents ancestral sequences at the genus level.
Recall vs. misclassification rates using leave-one-individual-out analysis for the order Charadriiformes (cytochrome oxidase 1 [COI] metabarcode) with paired-end 150 bp × 2 reads with 0% (A), 1% (B), and 2% (C) error/polymorphism, single-end 150 bp reads with 0% (D), 1% (E), and 2% (F) error/polymorphism, and single-end 300 bp reads with 0% (G), 1% (H), and 2% (I) error/polymorphism using kraken2, metaphlan2, MEGAN, pplacer, APPLES-2, and Tronko with cut-offs of 0, 5, 10, 15, and 20 using the Needleman–Wunsch alignment (solid line).
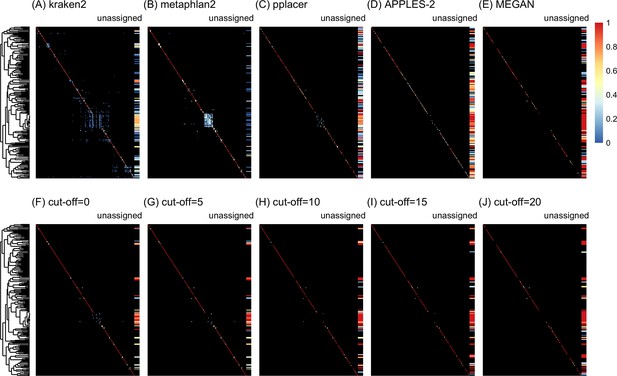
Confusion matrices at the species level of the order Charadriiformes using the leave-one-individual-out analysis with paired-end 150 bp × 2 reads with 2% error/polymorphism using kraken2 (A), metaphlan2 (B), pplacer (C), APPLES-2 (D), MEGAN (E), and Tronko using the Needleman–Wunsch alignment (NW) for cut-offs 0 (F), 5 (G), 10 (H), 15 (I), and (J) 20.
Unassigned column contains both unassigned queries and queries assigned to a lower taxonomic level. Phylogenetic tree represents ancestral sequences at the species level.
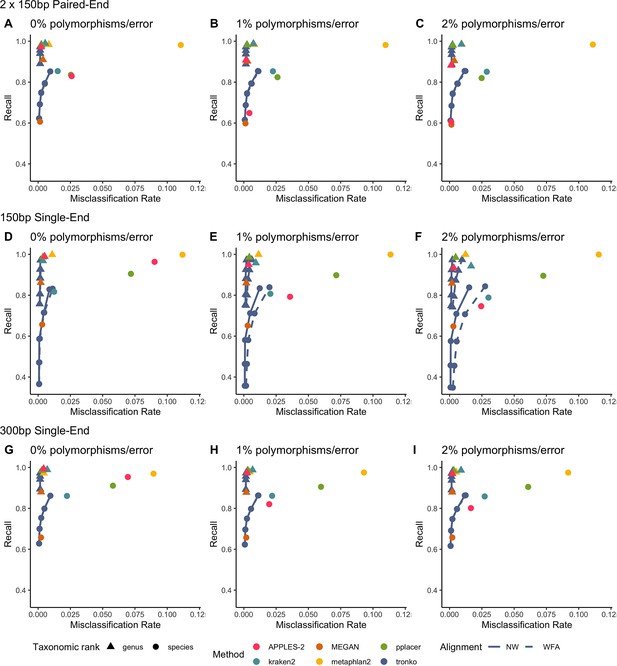
Recall vs. misclassification rates using leave-one-individual-out analysis for the order Charadriiformes (cytochrome oxidase 1 [COI] metabarcode) with paired-end 150 bp × 2 reads with 0% (A), 1% (B), and 2% (C) error/polymorphism, single-end 150 bp reads with 0% (D), 1% (E), and 2% (F) error/polymorphism, and single-end 300 bp reads with 0% (G), 1% (H), and 2% (I) error/polymorphism using kraken2, metaphlan2, MEGAN, pplacer, APPLES-2, and Tronko with cut-offs of 0, 5, 10, 15, and 20 using the Needleman–Wunsch alignment (solid line) and wavefront alignment (dashed line).
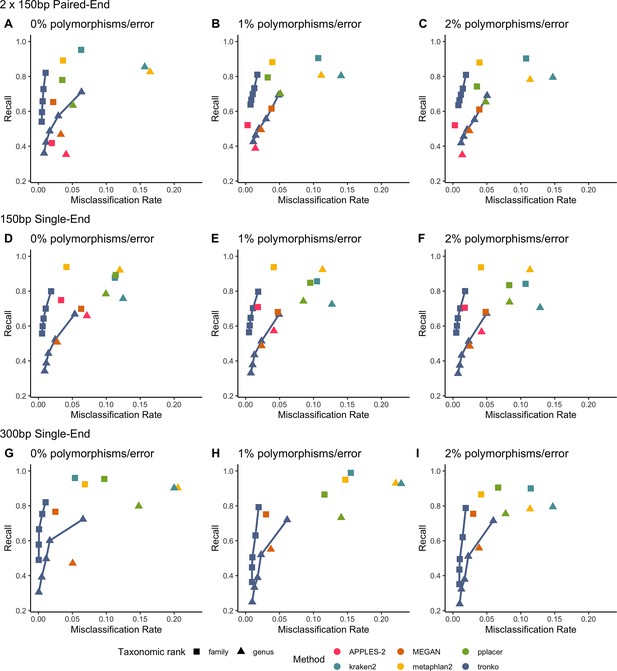
Recall vs. misclassification rates using leave-one-species-out analysis with bacteria species (16S metabarcode) with paired-end 150 bp × 2 reads with 0% (A), 1% (B), and 2% (C) error/polymorphism, single-end 150 bp reads with 0% (D), 1% (E), and 2% (F) error/polymorphism, and single-end 300 bp reads with 0% (G), 1% (H), and 2% (I) error/polymorphism using kraken2, metaphlan2, MEGAN, pplacer, APPLES-2 and Tronko with cut-offs of 0, 5, 10, 15, and 20 using the Needleman–Wunsch alignment (solid line).
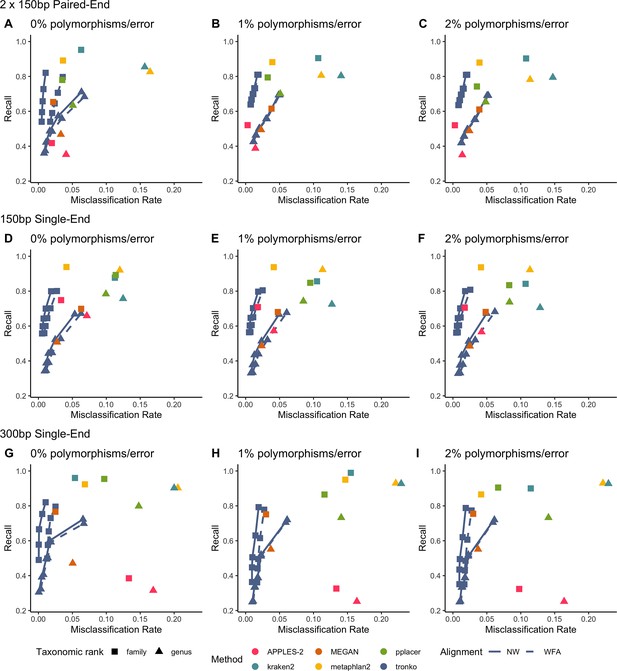
Recall vs. misclassification rates using leave-one-species-out analysis with bacteria species (16S metabarcode) with paired-end 150 bp × 2 reads with 0% (A), 1% (B), and 2% (C) error/polymorphism, single-end 150bp reads with 0% (D), 1% (E), and 2% (F) error/polymorphism, and single-end 300bp reads with 0% (G), 1% (H), and 2% (I) error/polymorphism using kraken2, metaphlan2, MEGAN, pplacer, APPLES-2, and Tronko with cut-offs of 0, 5, 10, 15, and 20 using the Needleman–Wunsch alignment (solid line) and wavefront alignment (dashed line).
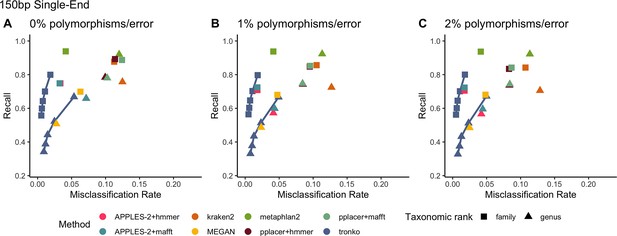
Recall vs. misclassification rates using leave-one-individual-out analysis for bacterial species (16S metabarcode) with paired-end 150 bp × 2 reads with 0% (A), 1% (B), and 2% (C) error/polymorphism using kraken2, metaphlan2, MEGAN, pplacer + hmmer, pplacer + mafft, APPLES-2 + hmmer, APPLES-2 + mafft, and Tronko with cut-offs of 0, 5, 10, 15, and 20 using the Needleman–Wunsch alignment (solid line).
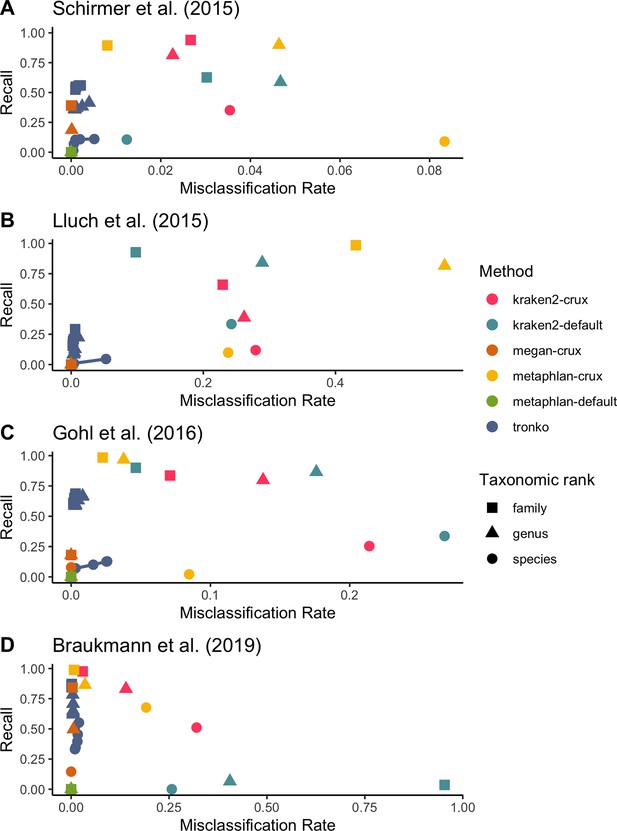
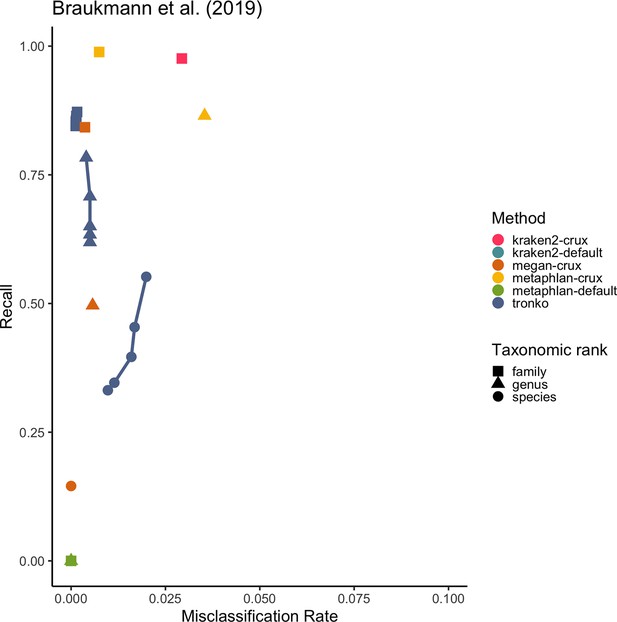
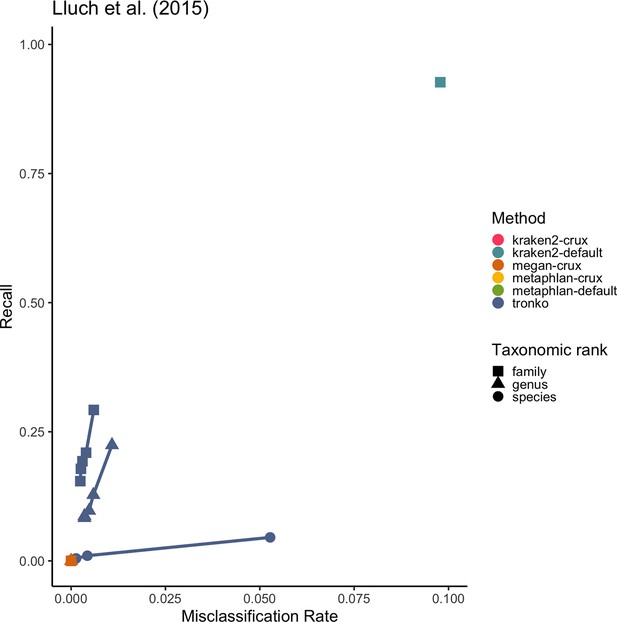
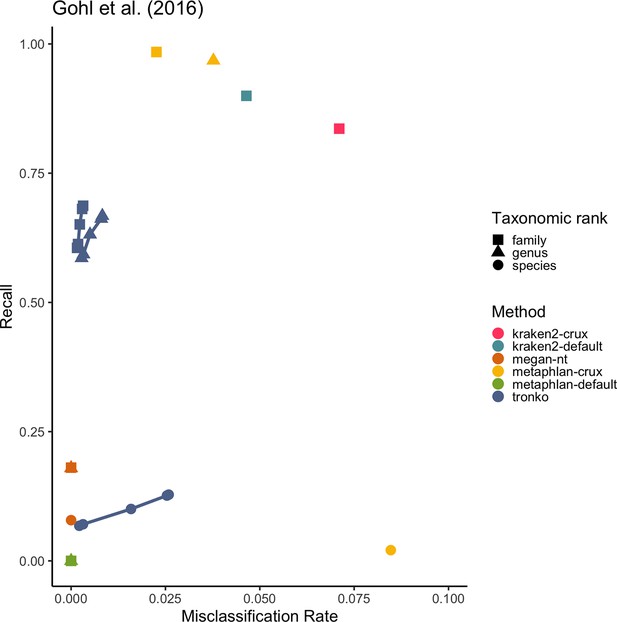
Recall vs. misclassification rates using mock communities from Schirmer et al., 2015 (A), Lluch et al., 2015 (B), Gohl et al., 2016 (C), and Braukmann et al., 2019 (D) using both Needleman–Wunsch and wavefront alignment algorithms.
Figures with smaller misclassification rates on the x-axis are available for Schirmer et al., 2015, Lluch et al., 2015, Gohl et al., 2016, Braukmann et al., 2019 in Figure 6—figure supplements 1, 2, and 4, respectively.
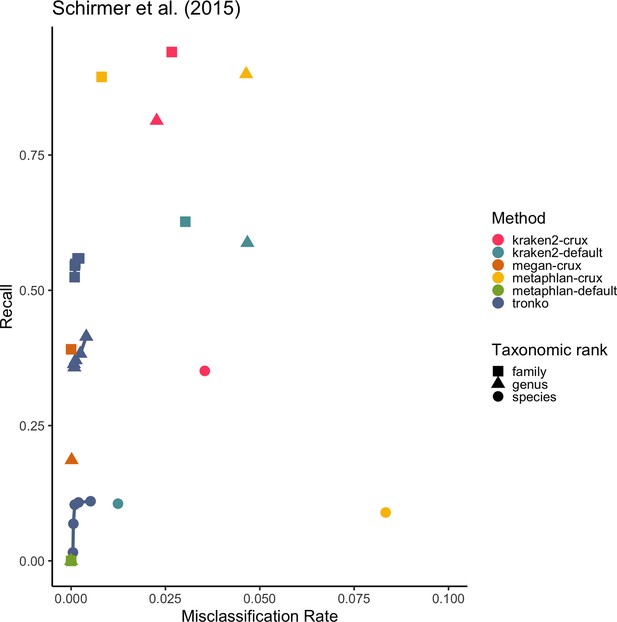
Close-up of Figure 6A.
Close-up of Figure 6A.
Close-up of Figure 6B.