ONC201/TIC10 enhances durability of mTOR inhibitor everolimus in metastatic ER+ breast cancer
Figures
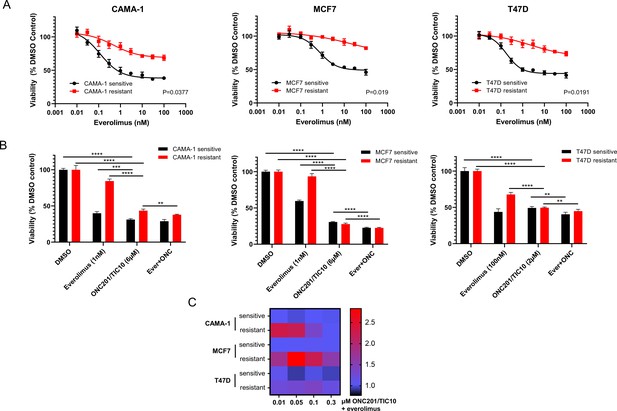
ONC201/TIC10 inhibits the proliferation of everolimus sensitive and resistant cells in 2D.
(A) Dose–response curves of CAMA-1, MCF7, and T47D everolimus sensitive and resistant cells under everolimus treatment. Cells were treated with increasing concentration of everolimus for 4 days and viability was measured using CellTiterGlo Chemiluminescent kit. (B) Cell viability after 4 days treatment with Dimethyl sulfoxide (DMSO), everolimus, ONC201/TIC10, or combination at the indicated concentrations. Data represent % viable cells compared with DMSO control treatment for each cell line and are shown as average of four replicates ± standard deviation (SD). **p < 0.01, ***p < 0.001, ****p < 0.0001. (C) Analysis of ONC201/TIC10 and everolimus interactions in 2D. Cells were treated with 1 nM everolimus (CAMA-1 and T47D) or 100 nM everolimus (MCF7), ONC201/TIC10, or combination at the indicated concentrations for 4 days in 2D and viability was measured using CellTiterGlo Chemiluminescent kit. The average Bliss Interaction Index was calculated and plotted as a heatmap in which red represents synergy and blue represents additivity.
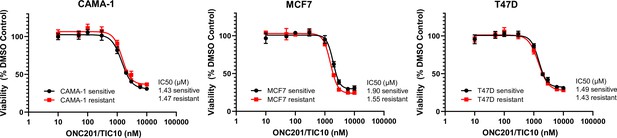
ONC201/TIC10 inhibits the proliferation of everolimus sensitive and resistant cells in 2D.
Dose–response curves of CAMA-1, MCF7, and T47D everolimus sensitive and resistant cells under ONC201/TIC10 treatment. Cells were treated with increasing concentration of ONC201/TIC10 for 4 days and viability was measured using CellTiterGlo Chemiluminescent kit. Data represent % viable cells compared with DMSO control treatment for each cell line and are shown as average of four replicates ± standard deviation (SD).
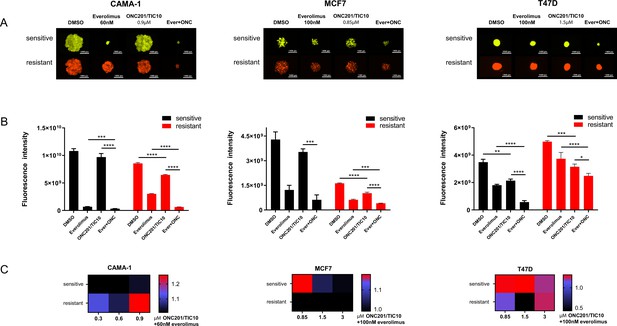
Combination of ONC201/TIC10 and everolimus inhibits spheroid growth in 3D.
(A) Representative images of spheroid growth of sensitive (Venus, green) or resistant (mCherry, red) cells cultured in everolimus, ONC201/TIC10, or combination treated media at the indicated concentrations for up to 18 days. (B) Fluorescence intensity of sensitive and resistant cells under various treatment conditions. Data are represented as average of three replicates ± standard deviation (SD). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (C) Analysis of ONC201/TIC10 and everolimus interactions in 3D. Spheroids were cultured in the presence of drug treatments for 18 days and fluorescence intensity measurements were captured. The average Bliss Interaction Index was calculated and plotted as a heatmap in which red represents synergy and blue represents additivity.
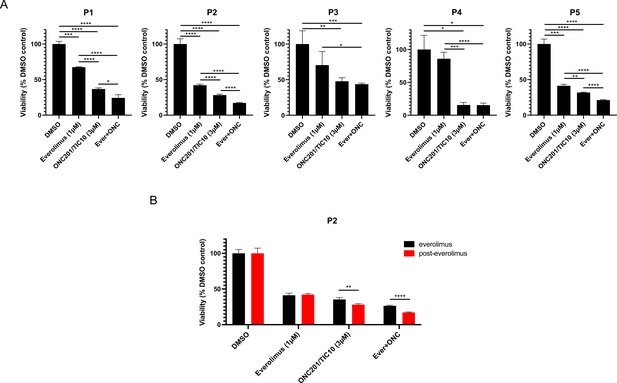
Combination therapy of ONC201/TIC10 and everolimus inhibits the growth of primary patient-derived cell spheroids.
(A) 3D cultures of primary patient-derived ER+ BC cells from ascites or pleural effusion treated with everolimus, ONC201/TIC10, or combination at the indicated concentrations for 4 days. Cell viability was measured using CellTiterGlo Chemiluminescent kit. (B) 3D cultures of primary patient-derived ER+ BC cells, while on everolimus treatment or post-everolimus treatment. Spheroids were treated with everolimus, ONC201/TIC10, or combination at the indicated concentrations for 4 days. Data represent % viable cells compared with DMSO control treatment and are shown as average of three replicates ± standard deviation (SD). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
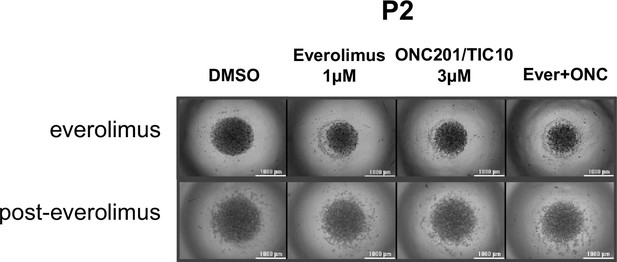
Combination therapy of ONC201/TIC10 and everolimus inhibits the growth of primary patient-derived cell spheroids.
Representative images of spheroid growth of primary patient-derived ER+ BC cells collected during everolimus treatment or post-everolimus and therapy resistant, cultured and treated with everolimus, ONC201/TIC10, or combination at the indicated concentrations for 4 days.
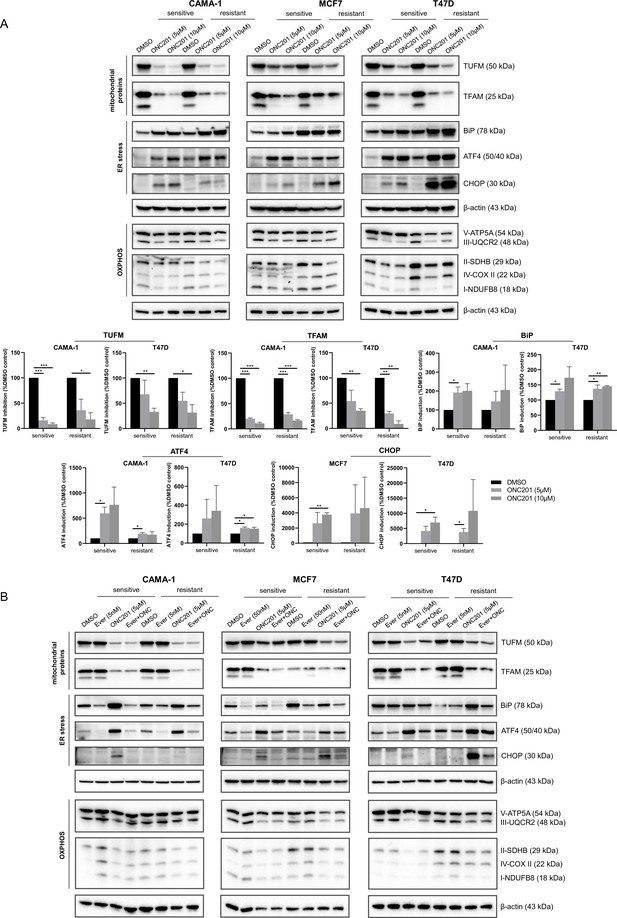
ONC201/TIC10 causes loss of mitochondrial proteins and activation of stress response in everolimus sensitive and resistant cells.
(A) CAMA-1, MCF7, T47D everolimus sensitive and resistant cells were treated for 24 hr with ONC201/TIC10 at the indicated concentrations, and cell lysates were immunoblotted for TUFM, TFAM, BiP, ATF4, CHOP, and OXPHOS complexes (Complex I subunit NDUFB8, Complex II subunit 30 kDa, Complex III subunit Core 2, Complex IV subunit II, and ATP synthase subunit alpha), and β-actin. Quantitation of TUFM, TFAM, BiP, ATF4, and CHOP using ImageJ analysis. Protein expression levels were normalized to β-actin and are shown as average of two replicates ± standard deviation (SD). *p < 0.05, **p < 0.01, ***p < 0.001. (B) CAMA-1, MCF7, T47D everolimus sensitive and resistant cells were treated for 24 hr with everolimus, ONC201/TIC10, and combination at the indicated concentrations, and cell lysates were immunoblotted for the same proteins as above.
-
Figure 4—source data 1
Original files of the full blot images for Figure 4A.
- https://cdn.elifesciences.org/articles/85898/elife-85898-fig4-data1-v1.zip
-
Figure 4—source data 2
Original files of the full blot images for Figure 4B.
- https://cdn.elifesciences.org/articles/85898/elife-85898-fig4-data2-v1.zip
-
Figure 4—source data 3
Figures with uncropped blot images for Figure 4A, B.
- https://cdn.elifesciences.org/articles/85898/elife-85898-fig4-data3-v1.pdf
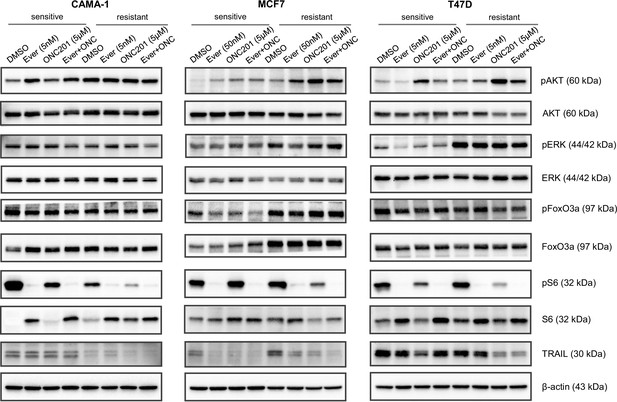
ONC201/TIC10 mechanism in everolimus sensitive and resistant cells is TRAIL independent.
CAMA-1, MCF7, T47D everolimus sensitive and resistant cells were treated for 24 hr with everolimus, ONC201/TIC10, and combination at the indicated concentrations. Cell lysates were immunoblotted for pAKT, AKT, pERK, ERK, pFoxO3a, FoxO3a, TRAIL, pS6, S6, and β-actin.
-
Figure 4—figure supplement 1—source data 1
Original files of the full blot images for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/85898/elife-85898-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Figures with uncropped blot images for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/85898/elife-85898-fig4-figsupp1-data2-v1.pdf
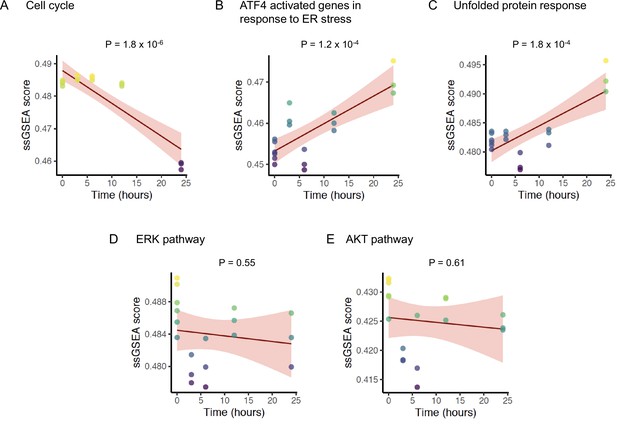
Change in pathway activity over time in response to ONC201/TIC10.
Scatter plots displaying the enrichment scores (Y-axis) of (A) REACTOME cell cycle signature, (B) REACTOME ATF4 activated genes in response to endoplasmic reticulum stress signature, (C) REACTOME unfolded protein response UPR signature, (D) BIOCARTA ERK pathway signature, and (E) BIOCARTA AKT pathway signature over time (X-axis). The solid lines and shaded area indicate linear fit and 95% confidence intervals, respectively, with p-value of the fit indicated above each plot. The analysis includes data from six replicates at time 0 hr, and three replicates each at time 3, 6, 12, and 24 hr for a total n = 18.
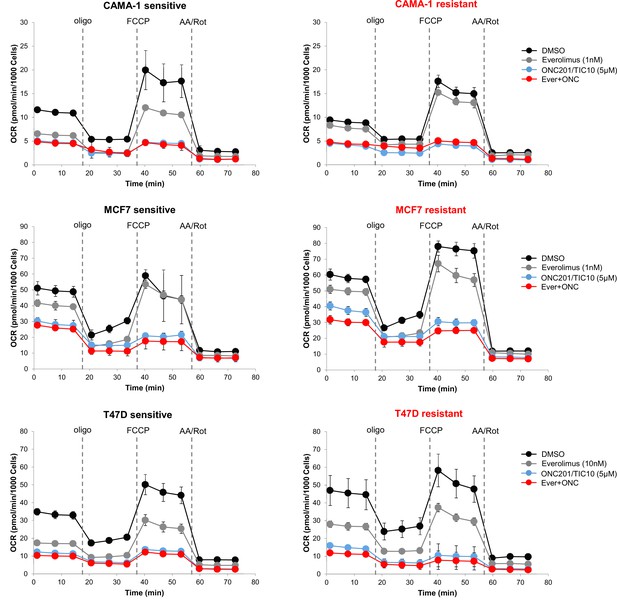
ONC201/TIC10 inhibits mitochondrial respiration in everolimus sensitive and resistant cells.
Cells were treated for 18 hr with indicated concentrations of everolimus, ONC201/TIC10, and combination. Mitochondrial respiration was measured using Seahorse XF Cell Mito Stress assay and oxygen consumption rates (OCR) are shown. Values were normalized to cell number generated from fluorescence intensity measurements and are represented as average of three replicates.
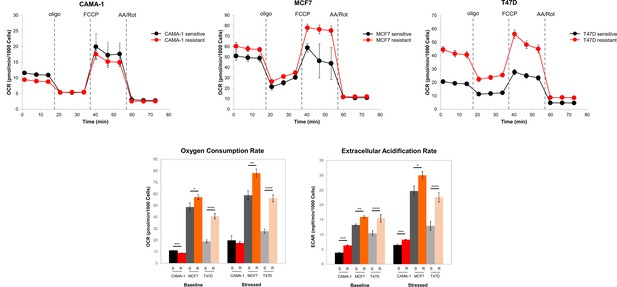
Mitochondrial respiration in everolimus sensitive and resistant cells.
Cells were cultured for 18 hr. Mitochondrial respiration was measured using Seahorse XF Cell Mito Stress assay and oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) graphs are shown. Values were normalized to cell number generated from fluorescence intensity measurements and are represented as average of three replicates ± standard deviation (SD). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
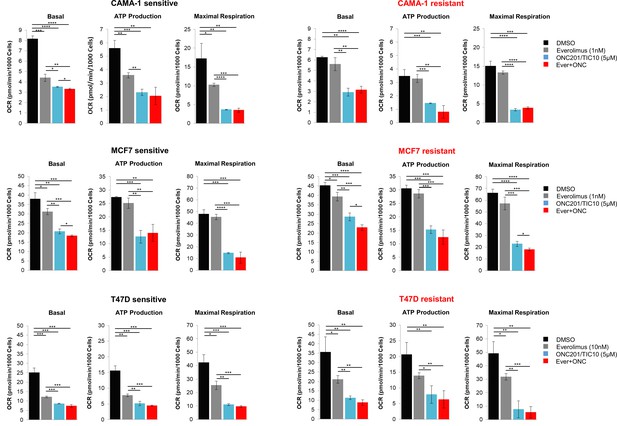
ONC201/TIC10 inhibits mitochondrial respiration in everolimus sensitive and resistant cells.
Cells were treated for 18 hr with indicated concentrations of ONC201/TIC10 and everolimus. Mitochondrial respiration was measured using Seahorse XF Cell Mito Stress assay and oxygen consumption rates (OCR) bar graphs are shown for basal, maximal respiration and ATP production. Values were normalized to cell number generated from fluorescence intensity measurements and are represented as average of three replicates ± standard deviation (SD). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
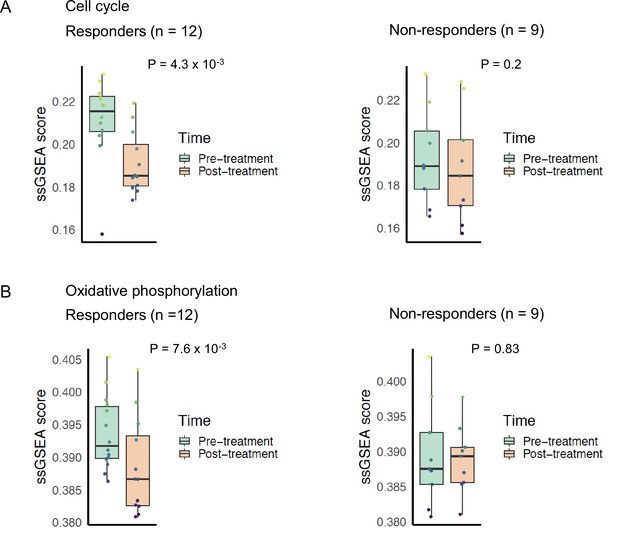
Cell cycle and oxidative phosphorylation pathway activity and neoadjuvant everolimus response.
Box plots comparing cell cycle pathway activity indicated by (A) REACTOME cell cycle signature enrichment scores and (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) oxidative phosphorylation signature enrichment scores between pre- and post-treatment samples. The left panel shows patients that were classified as responders to everolimus, while the right panel shows patients that were classified as non-responders. Colored boxes indicate interquartile range, horizontal bars indicate median, and the whiskers indicate first and third quartiles. p-values from paired two-tailed t-test comparing pre-treatment vs. post-treatment scores are indicated above the plots.
Tables
Treatment history of patients included in the study.
| Patient diagnosis | Sample | Therapy lines | Drugs | Therapy lines post-everolimus | |
|---|---|---|---|---|---|
| P1 | Metastatic ER+/HER2− | Ascites | 6 | 12 | 2 |
| P2 | Metastatic ER+/PR+/HER2− | Ascites | 7 | 9 | 2 |
| P3 | Metastatic ER+/PR+/HER2− | Pleural effusion | 7 | 9 | 1 |
| P4 | Metastatic ER+/PR+/HER2− | Ascites | 10 | 14 | 5 |
| P5 | Metastatic ER+/PR+/HER2− | Ascites | 5 | 6 | 1 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo-sapiens) female | CAMA-1 Breast; Mammary gland: adenocarcinoma | ATCC | HTB-21 | |
| Cell line (Homo-sapiens) female | MCF7 Breast; Mammary gland: adenocarcinoma epithelial cell | ATCC | HTB-22 | |
| Cell line (Homo-sapiens) female | T47D Breast; Mammary gland: carcinoma ductal epithelial cell | ATCC | HTB-133 | |
| Cell line (Homo-sapiens) female | Primary, P1, P2, P3, P4, P5 | This paper. City of Hope IRB #07047 and #17334 | P1, P2, P3, P4, P5 | Pleural effusion or ascites samples from patients |
| Transfected construct (mammalian) | LeGO-V2 (Venus) | Weber et al., 2008 Addgene #27340 | Addgene #27340; RRID:Addgene_27340 | Lentiviral construct to transfect and express Venus fluorescent protein |
| Transfected construct (mammalian) | LeGO-C2 (mCherry) | Weber et al., 2008 Addgene #27339 | Addgene #27339; RRID:Addgene_27339 | Lentiviral construct to transfect and express mCherry fluorescent protein |
| Antibody | anti-BiP (C50B12) (rabbit monoclonal) | Cell Signaling Technology | Cat#3177; RRID:AB_2119845 | WB (1:1000) |
| Antibody | anti-pFoxO3a (Ser294) (rabbit polyclonal) | Cell Signaling Technology | Cat#5538; RRID:AB_10696878 | WB (1:1000) |
| Antibody | anti-FoxO3a (D19A7) (rabbit monoclonal) | Cell Signaling Technology | Cat#12829; RRID:AB_2636990 | WB (1:1000) |
| Antibody | anti-pERK1/2 Thr202/Tyr204 (197G2) (rabbit monoclonal) | Cell Signaling Technology | Cat#4377; RRID:AB_331775 | WB (1:1000) |
| Antibody | anti-ERK1/2 (137F5) (rabbit monoclonal) | Cell Signaling Technology | Cat#4695; RRID:AB_390779 | WB (1:1000) |
| Antibody | anti-pAKT Ser473 (D9E) (rabbit monoclonal) | Cell Signaling Technology | Cat#4060; RRID:AB_2315049 | WB (1:1000) |
| Antibody | anti-AKT (C67E7) (rabbit monoclonal) | Cell Signaling Technology | Cat#4691; RRID:AB_915783 | WB (1:1000) |
| Antibody | anti-pS6 Ser240/244 Ribosomal Protein (D68F8) (rabbit monoclonal) | Cell Signaling Technology | Cat#5364; RRID:AB_10694233 | WB (1:1000) |
| Antibody | anti-S6 Ribosomal Protein (54D2) (mouse monoclonal) | Cell Signaling Technology | Cat#2317; RRID:AB_2238583 | WB (1:1000) |
| Antibody | anti-β-actin (8H10D10) (mouse monoclonal) (HRP Conjugate) | Cell Signaling Technology | Cat#12262; RRID:AB_2566811 | WB (1:2000) |
| Antibody | anti-β-actin (13E5) (rabbit monoclonal) (HRP Conjugate) | Cell Signaling Technology | Cat#5125; RRID:AB_1903890 | WB (1:2000) |
| Antibody | anti-rabbit IgG, HRP-linked (goat polyclonal) | Cell Signaling Technology | Cat#7074; RRID:AB_2099233 | WB (1:2000) |
| Antibody | anti-mouse IgG, HRP-linked (horse polyclonal) | Cell Signaling Technology | Cat#7076; RRID:AB_330924 | WB (1:2000) |
| Antibody | anti-CREB-2/ATF-4 (B-3) (mouse monoclonal) | Santa Cruz Biotechnology | Cat#390063; RRID:AB_2810998 | WB (1:300) |
| Antibody | anti-mtTFA (TFAM) (C-9) (mouse monoclonal) | Santa Cruz Biotechnology | Cat#376672; RRID:AB_11150497 | WB (1:200) |
| Antibody | anti-TRAIL (55B709.3) (mouse monoclonal) | Thermo Fisher Scientific | Cat# MA1-41027; RRID:AB_1087999 | WB (1.5 μg/ml) |
| Antibody | anti-CHOP (rabbit polyclonal) | Proteintech | Cat#15204-1-AP; RRID:AB_2292610 | WB (1:1000) |
| Antibody | anti-TUFM (rabbit polyclonal) | Thermo Fisher Scientific | Cat#PA5-27511; RRID:AB_2544987 | WB (1:500) |
| Antibody | anti-Total OXPHOS (mouse monoclonal) | Abcam | Cat#ab110411; RRID:AB_2756818 | WB (1:1000) |
| Chemical compound, drug | Everolimus (RAD001) | Selleckchem | Cat#S1120 | Dissolved in DMSO |
| Chemical compound, drug | ONC201/TIC10 | Selleckchem | Cat#S7963 | Dissolved in DMSO |
| Commercial assay or kit | CellTiter-Glo Luminescent Cell Viability Assay | Promega | Cat#G7573 | |
| Commercial assay or kit | CellTiter-Glo 3D Cell Viability Assay | Promega | Cat#G9682 | |
| Commercial assay or kit | EasySep CD45 Depletion Kit II | StemCell Technologies | Cat#17898 | |
| Commercial assay or kit | EasySep Dead Cell Removal (Annexin V) Kit | StemCell Technologies | Cat#17899 | |
| Commercial assay or kit | Agilent Seahorse XF Cell Mito Stress Test Kit | Agilent Technologies | Cat#103015-100 | |
| Software | GraphPad Prism software | GraphPad (https://graphpad.com) | RRID:SCR_002798 | Version 9.3.1 |
| Software | Gen5 software | Biotek Instruments (https://www.agilent.com/) | Version 3.05 | |
| Software | Seahorse Wave Desktop Software 2.6 | Agilent Technologies (https://www.agilent.com/) | RRID:SCR_014526 | Version 2.6 |
| Software | ImageJ software | ImageJ (http://imagej.nih.gov/ij/) | RRID:SCR_003070 |





