Structural insights into regulation of CNNM-TRPM7 divalent cation uptake by the small GTPase ARL15
Figures
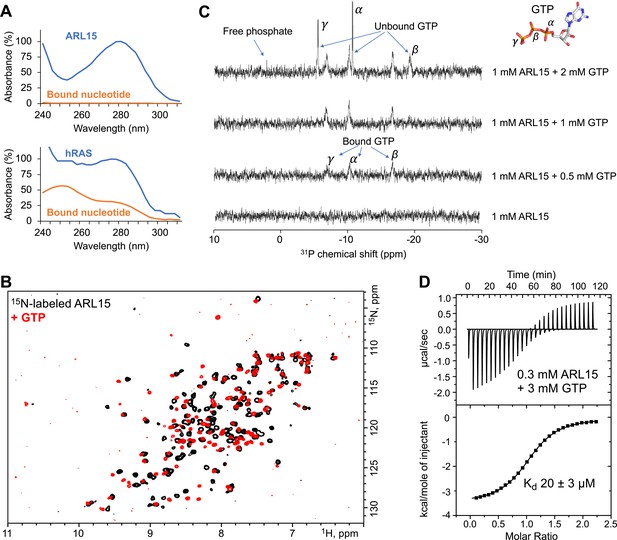
ADP-ribosylation factor-like GTPase 15 (ARL15) (32–197) purifies without nucleotide bound and has weak affinity for GTP.
(A) UV spectra of recombinant ARL15 and the GTPase hRAS. Spectra were recorded before (blue) and after heat treatment (orange) to precipitate the protein component. Unlike hRAS, ARL15 has no nucleotide bound. (B) 1H-15N correlation NMR spectra of 100 µM ARL15 unliganded (black) and with 500 µM GTP (red). (C) Phosphorus (31P) NMR spectra of recombinant ARL15 titrated with GTP. Prior to addition of GTP, no signals are visible. GTP binds until ARL15 is saturated whereupon signals for both bound and free GTP are observed. (D) Isothermal titration calorimetry shows ARL15 binds GTP with mid-micromolar affinity.
-
Figure 1—source data 1
Fitting parameters for isothermal titration calorimetry (ITC) thermogram of ADP-ribosylation factor-like GTPase 15 (ARL15) binding to GTP in Figure 1D.
- https://cdn.elifesciences.org/articles/86129/elife-86129-fig1-data1-v1.pdf
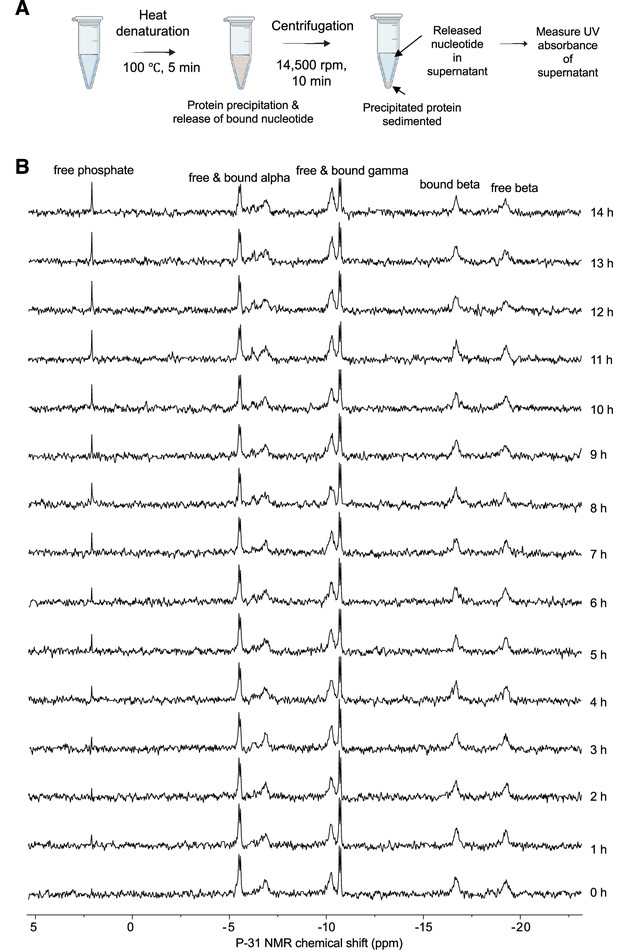
ADP-ribosylation factor-like GTPase 15 (ARL15) has low intrinsic GTPase activity.
(A) Procedure to detect bound nucleotide. Figure prepared with BioRender. (B) Measurement of intrinsic GTPase activity. 31P NMR spectra of 1 mM ARL15 with 2 mM GTP collected overnight at 25°C show an accumulation of free phosphate and a small (~25%) decrease in free GTP. The loss corresponds to a turnover rate of 0.000005 s–1. Notably, there is no evidence of formation of GDP, which would appear as a decrease in the β signal relative to the α and γ signals.
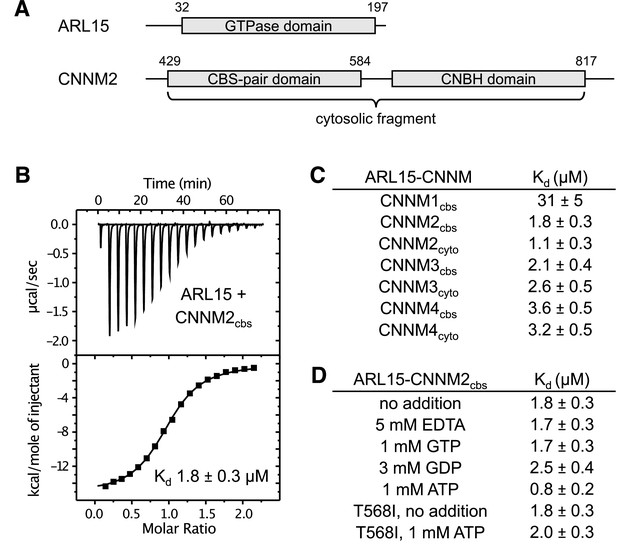
ADP-ribosylation factor-like GTPase 15 (ARL15) binds cystathionine-β-synthase-pair domain divalent metal cation transport mediator (CNNM) cystathionine-β-synthase (CBS)-pair domains.
(A) Domains of ARL15 and CNNM2 proteins used in binding experiments. The four CNNM protein have the same domain organization. (B) Isothermal titration calorimetry experiment between the ARL15 GTPase domain and CNNM2 CBS-pair domain. (C) Binding affinities (Kd) between ARL15 GTPase domain and CBS-pair and cytosolic fragments of CNNM proteins measured by isothermal titration calorimetry (ITC). (D) Effect of nucleotides on the affinity of the ARL15-CNNM2 interaction measured by ITC. The CNNM2 T568I mutant, which is unable to bind ATP, bound ARL15 with the same affinity as the wild-type CNNM2 CBS-pair domain but did not show increased affinity upon addition of ATP.
-
Figure 2—source data 1
Isothermal titration calorimetry (ITC) thermograms showing binding between ADP-ribosylation factor-like GTPase 15 (ARL15) GTPase domain and cystathionine-β-synthase (CBS)-pair and cytosolic fragments of cystathionine-β-synthase-pair domain divalent metal cation transport mediator (CNNM) proteins in Figure 1B and C.
- https://cdn.elifesciences.org/articles/86129/elife-86129-fig2-data1-v1.pdf
-
Figure 2—source data 2
Isothermal titration calorimetry (ITC) thermograms showing binding between ADP-ribosylation factor-like GTPase 15 (ARL15) GTPase domain and cystathionine-β-synthase (CBS)-pair domain of CBS-pair domain divalent metal cation transport mediator 2 (CNNM2) in presence of 5 mM EDTA, 1 mM GTP, and 3 mM GDP in Figure 1D.
- https://cdn.elifesciences.org/articles/86129/elife-86129-fig2-data2-v1.pdf
-
Figure 2—source data 3
Isothermal titration calorimetry (ITC) thermograms showing binding between ADP-ribosylation factor-like GTPase 15 (ARL15) GTPase domain and cystathionine-β-synthase (CBS)-pair domain of CBS domain divalent metal cation transport mediator 2 (CNNM2) (wild-type [WT] and T568I mutant) in presence or absence of Mg-ATP in Figure 1D.
- https://cdn.elifesciences.org/articles/86129/elife-86129-fig2-data3-v1.pdf
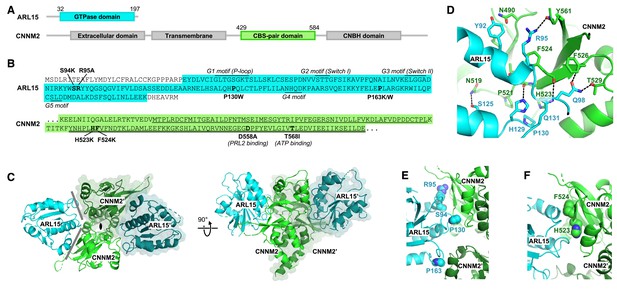
Crystal structure of ADP-ribosylation factor-like GTPase 15 (ARL15) bound to the cystathionine-β-synthase (CBS)-pair domain of CBS-pair domain divalent metal cation transport mediator 2 (CNNM2).
(A) Domain organization of the proteins. (B) Sequences of the fragments crystallized and residues mutated. The typical GTPase motifs are indicated for ARL15. Within the CNNM2 CBS-pair domain, the CBS1 and CBS2 motifs are singly and doubly underlined. (C) Crystal structure of two ARL15 molecules bound to a CBS-pair domain dimer. There are two distinct interfaces (gray lines) between the ARL15 and CBS-pair monomers. (D) Detail of intermolecular contacts between ARL15 and the CNNM2 CBS-pair domain. (E) Interfacial ARL15 residues tested by mutagenesis. (F) Interfacial CNNM2 residues tested by mutagenesis.
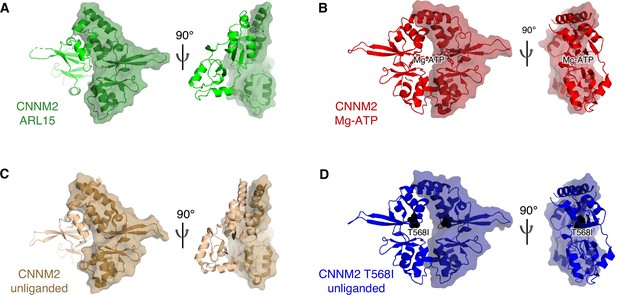
Structural comparison of the cystathionine-β-synthase-pair domain divalent metal cation transport mediator 2 (CNNM2) cystathionine-β-synthase (CBS)-pair domain dimerization when (A) bound to ADP-ribosylation factor-like GTPase 15 (ARL15) (green), (B) bound to Mg-ATP (red, PDB code 4P1O), (C) no ligand bound (brown, 4IYS), and (D) T568I mutant (blue, 4IY4).
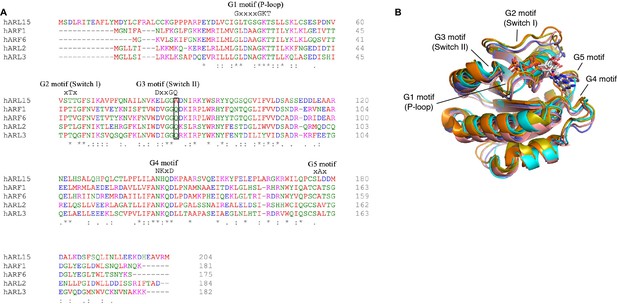
Comparison of ADP-ribosylation factor-like GTPase 15 (ARL15) to related small GTPases.
(A) Sequence alignment of human ARL15 and other ARL/ARF GTPases. The glutamine residue in the G3 motif that is alanine in ARL15 is boxed. (B) Structural comparison of ARL15 (cyan) to small GTPases: ARL2 (pink, PDB code 4GOK), ARL3 (orange, 3BH6), ARF1 (olive, 1O3Y), ARF6 (violet, 4KAX).
Crystal structure of two ADP-ribosylation factor-like GTPase 15 (ARL15) molecules (cyan and teal) bound to a cystathionine-β-synthase (CBS)-pair domain dimer (light and dark green).
Residues tested by mutagenesis are indicated.
Comparison of ADP-ribosylation factor-like GTPase 15 (ARL15) structure (cyan) and the related small GTPases: ARL2 (pink, PDB code 4GOK), ARL3 (orange, 3BH6), ARF1 (olive, 1O3Y), ARF6 (violet, 4KAX).
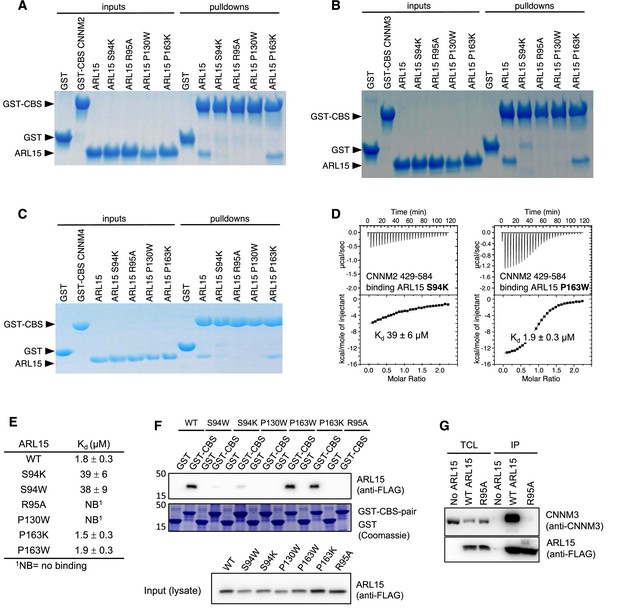
Mutagenesis of ADP-ribosylation factor-like GTPase 15 (ARL15) confirms the cystathionine-β-synthase-pair domain divalent metal cation transport mediator (CNNM)-binding site.
Pulldowns of recombinant WT and mutant ARL15 GTPase domains (residues 32–197) by (A) GST-CNNM2 cystathionine-β-synthase (CBS)-pair domain (residues 429–584), (B) GST-CNNM3 CBS-pair domain (residues 299–452), (C) GST-CNNM4 CBS-pair domain (residues 356–511). (D) Isothermal titration of mutants of ARL15 GTPase domain and CNNM2 CBS-pair domain. (E) CNNM2 binding affinities (Kd) of ARL15 GTPase mutants measured by isothermal titration calorimetry (ITC). (F) Pulldown of wild-type ARL15 (WT) and the indicated mutants expressed in HEK-293T cells by GST-fused CNNM2 CBS-pair domain. (G) Co-immunoprecipitation of FLAG-tagged ARL15 with native CNNM3 shows that the ARL15 R95A mutation fully blocks binding. No ARL15, untransfected; WT, wild-type.
-
Figure 4—source data 1
Uncropped gels of pulldowns of recombinant wild-type (WT) and mutant ADP-ribosylation factor-like GTPase 15 (ARL15) GTPase domains (residues 32–197) by GST-cystathionine-β-synthase-pair domain divalent metal cation transport mediator 2 (CNNM2) cystathionine-β-synthase (CBS)-pair domain (residues 429–584) in Figure 4A–C.
- https://cdn.elifesciences.org/articles/86129/elife-86129-fig4-data1-v1.pdf
-
Figure 4—source data 2
Isothermal titration calorimetry (ITC) thermograms of cystathionine-β-synthase-pair domain divalent metal cation transport mediator 2 (CNNM2) cystathionine-β-synthase (CBS)-pair domain binding mutants of ADP-ribosylation factor-like GTPase 15 (ARL15) GTPase domain (S94K, S94W, R95A, P130W, P163K, and P163W) in Figure 4D and E.
- https://cdn.elifesciences.org/articles/86129/elife-86129-fig4-data2-v1.pdf
-
Figure 4—source data 3
Uncropped blots and gels of pulldown of wild-type (WT) ADP-ribosylation factor-like GTPase 15 (ARL15) (WT) and the indicated mutants expressed in HEK-293T cells by GST-fused cystathionine-β-synthase-pair domain divalent metal cation transport mediator 2 (CNNM2) cystathionine-β-synthase (CBS)-pair domain in Figure 4F.
- https://cdn.elifesciences.org/articles/86129/elife-86129-fig4-data3-v1.pdf
-
Figure 4—source data 4
Uncropped blots images of co-immunoprecipitation of FLAG-tagged ADP-ribosylation factor-like GTPase 15 (ARL15) with native cystathionine-β-synthase-pair domain divalent metal cation transport mediator 3 (CNNM3) show the R95A mutation fully blocks binding in Figure 4G.
- https://cdn.elifesciences.org/articles/86129/elife-86129-fig4-data4-v1.pdf
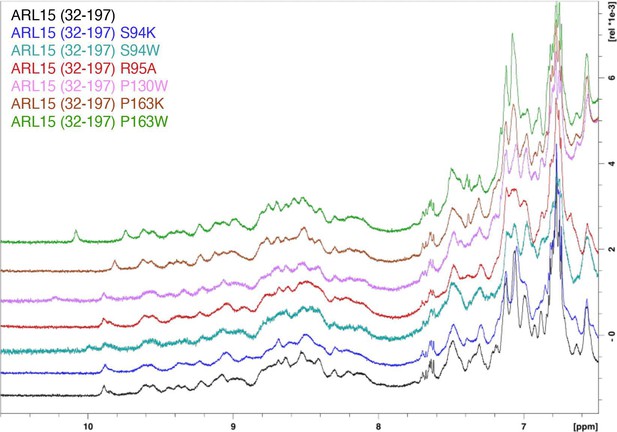
ADP-ribosylation factor-like GTPase 15 (ARL15) mutants are properly folded.
Downfield 1H NMR spectra of ARL15 GTPase domain (residues 32–197) and mutants (S94K, S94W, R95A, P130W, P163K, P163W).
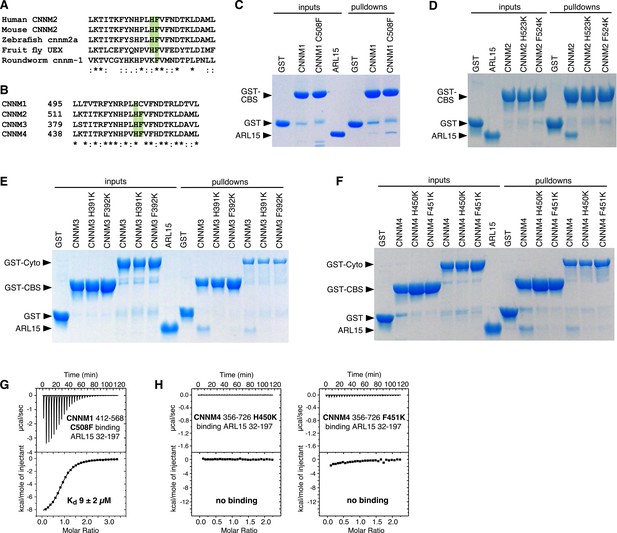
Mutagenesis of cystathionine-β-synthase-pair domain divalent metal cation transport mediator (CNNM) confirms conservation of ADP-ribosylation factor-like GTPase 15 (ARL15)-binding site.
(A) Conservation of key ARL15-binding residues (green) in different species. (B) Conservation of ARL15-binding residues in human CNNMs. (C–F) Pulldown of recombinant GTPase domain (residues 32–197) by GST fusions with (C) CNNM1 (residues 412–568), (D) CNNM2 (residues 429–584), (E) CNNM3 (residues 299–452 and residues 299–658), (F) CNNM4 (residues 356–511 and residues 356–726). (G) Isothermal titration calorimetry (ITC) thermogram of ARL15 GTPase domain and CNNM1 cystathionine-β-synthase (CBS)-pair domain with C508F mutation shows a threefold improvement in affinity. (H) Thermograms of CNNM4 cytosolic fragment H450K and F451K mutants show no ARL15 binding.
-
Figure 5—source data 1
Uncropped gels of pulldown of recombinant GTPase domain (residues 32–197) by wild-type (WT) and mutant GST-fused cystathionine-β-synthase-pair domain divalent metal cation transport mediator (CNNM) 1–4 in Figure 5C–F.
- https://cdn.elifesciences.org/articles/86129/elife-86129-fig5-data1-v1.pdf
-
Figure 5—source data 2
Isothermal titration calorimetry (ITC) thermograms of ADP-ribosylation factor-like GTPase 15 (ARL15) GTPase domain binding mutants of cystathionine-β-synthase-pair domain divalent metal cation transport mediator (CNNM) 1–4 cystathionine-β-synthase (CBS)-pair domains and CNNM4 cytosolic domain in Figure 5G and H.
- https://cdn.elifesciences.org/articles/86129/elife-86129-fig5-data2-v1.pdf
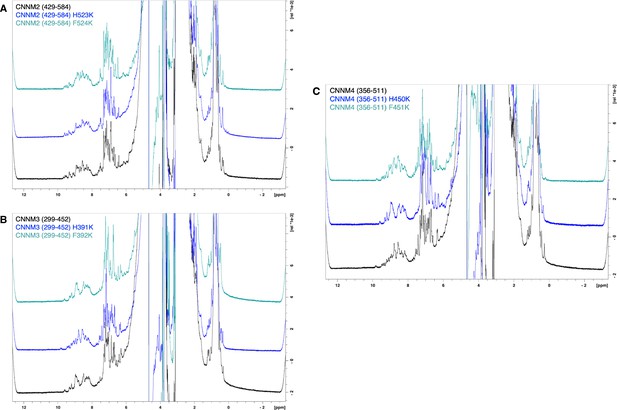
Cystathionine-β-synthase-pair domain divalent metal cation transport mediator 2 (CNNM2) cystathionine-β-synthase (CBS)-pair domain mutants are properly folded.
1H NMR spectra of (A) CNNM2 WT, H523K, and F524K mutants, (B) CNNM3 WT, H391K, and F392K mutants, and (C) CNNM4 WT, H450K, and F451K mutants.
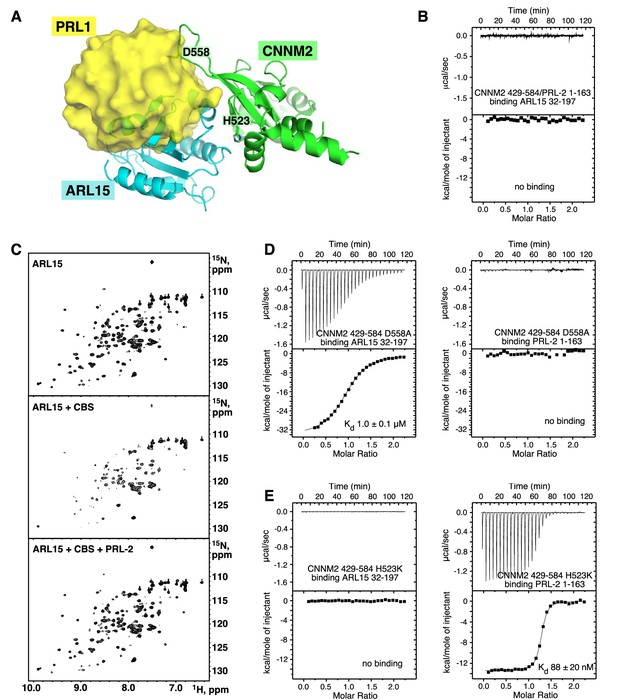
ADP-ribosylation factor-like GTPase 15 (ARL15) and phosphatases of regenerating liver (PRLs) have overlapping but distinct binding sites.
(A) Overlay of the structures of the cystathionine-β-synthase-pair domain divalent metal cation transport mediator 2 (CNNM2) complexes with ARL15 (cyan) and PRL1 (yellow, PDB code 6WUS) shows that simultaneous binding is not possible. (B) Isothermal titration calorimetry (ITC) thermogram shows no binding of ARL15 to the preformed complex of CNNM2 and PRL2. (C) 1H-15N correlation NMR spectra of 100 µM 15N-labeled ARL15 alone (top), in the presence of 100 µM CNNM2 cystathionine-β-synthase (CBS)-pair domain (middle), and after addition of 150 µM PRL2 (bottom). The ARL15 NMR signals are attenuated upon binding CNNM2 but reappear when ARL15 is displaced by PRL2. (D) ITC thermograms demonstrating that the CNNM2 D558A mutation specifically disrupts PRL2 binding. (E) ITC thermograms demonstrating that the CNNM2 H523K mutation specifically disrupts ARL15 binding.
-
Figure 6—source data 1
Isothermal titration calorimetry (ITC) experiments showing ADP-ribosylation factor-like GTPase 15 (ARL15) and phosphatases of regenerating liver (PRLs) have overlapping but distinct cystathionine-β-synthase-pair domain divalent metal cation transport mediator 2 (CNNM2)-binding sites in Figure 6B, D, and E.
- https://cdn.elifesciences.org/articles/86129/elife-86129-fig6-data1-v1.pdf
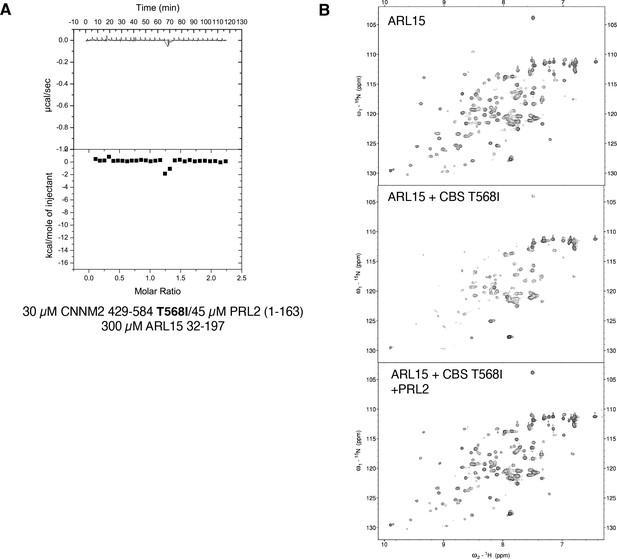
Competition between ADP-ribosylation factor-like GTPase 15 (ARL15) and phosphatase of regenerating liver 2 (PRL2) binding to cystathionine-β-synthase-pair domain divalent metal cation transport mediator 2 (CNNM2) is not affected by the CNNM2 T568I mutation.
(A) Isothermal titration calorimetry (ITC) thermogram shows no binding of ARL15 to the preformed complex of CNNM2 T568I and PRL2. (B) 1H-15N correlation NMR spectra of 100 µM 15N-labeled ARL15 alone (top), in the presence of 100 µM CNNM2 cystathionine-β-synthase (CBS)-pair domain mutant T568I (middle), and after addition of 150 µM PRL2 (bottom). The ARL15 NMR signals are attenuated upon binding the CNNM2 T568I mutant but reappear when ARL15 is displaced by PRL2.
Superposition of crystal structures of cystathionine-β-synthase-pair domain divalent metal cation transport mediator 2 (CNNM2) cystathionine-β-synthase (CBS)-pair domain (green) bound to ADP-ribosylation factor-like GTPase 15 (ARL15) (cyan) and phosphatase of regenerating liver 1 (PRL1) (yellow, PDB code 6WUS).
CNNM2 residues His523 and Asp558 are essential for binding respectively ARL15 and PRL1.

Disruption of the cystathionine-β-synthase-pair domain divalent metal cation transport mediator (CNNM)-binding site blocks ADP-ribosylation factor-like GTPase 15 (ARL15) regulation of CNNM and transient receptor potential ion channel subfamily M member 7 (TRPM7) divalent cation transport.
(A) CNNM2 Mg2+ efflux is inhibited by ARL15 but not the R95A mutant. HEK-293T cells transfected with the indicated constructs were loaded with a magnesium sensitive dye and fluorescence monitored following Mg2+ depletion. The mean relative fluorescence intensities of 10 cells are shown. (B) Representative TRPM7 whole-cell currents from 293-TRPM7 cells from untransfected controls (no ARL15), wild-type, and R95A mutant ARL15 transfectants. (C) Average current density of the different groups from (B). (D) Zinc influx assay using the FluoZin-3 Zn2+ indicator was used to monitor TRPM7 function in untransfected cells or transfected with wild-type or mutant ARL15. Images shown are taken at a time point 5–10 min after application of 30 μM ZnCl2 to stimulate Zn2+ influx. (E) Cell counts of fluorescence intensity from (D). A total of 50 cells per condition were randomly selected for quantification. *** indicates a p-value of less than 0.0001.
-
Figure 7—source data 1
Statistical analysis of the decrease in fluorescence in Figure 7A.
The fluorescence for 10 cells in each condition was measured in the first and last frames of the 5 min efflux assay and the ratio calculated. The significance between conditions was evaluated via Wilcoxon analysis. The statistical analysis was performed in R suite, and the figure prepared using the ggplot2 extension package.
- https://cdn.elifesciences.org/articles/86129/elife-86129-fig7-data1-v1.pdf
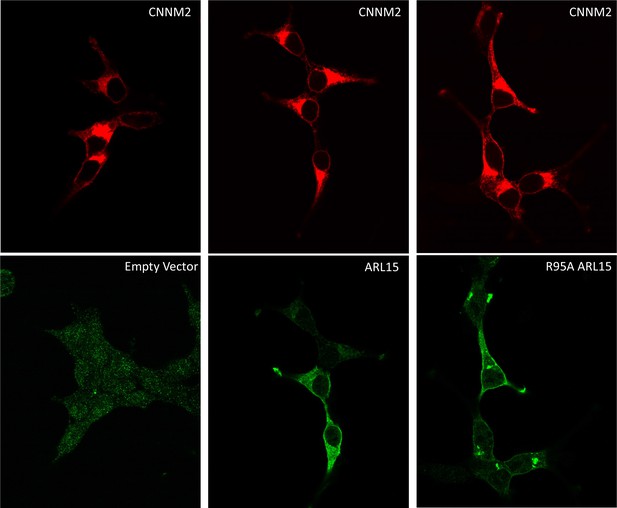
Localization of HEK239T cells transfected with mCNNM2-mCherry and FLAG-ADP-ribosylation factor-like GTPase 15 (ARL15).
Confocal microscopy shows that CNNM2 localizes to the plasma membrane and an intracellular compartment, possibly the Golgi apparatus. This localization is not affected by ARL15 expression. Both wild-type ARL15 and the R95A mutant localize to the plasma membrane as well as internal structures.
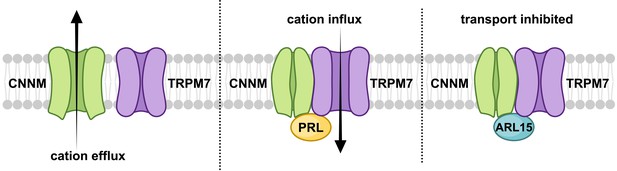
Model of regulation of cystathionine-β-synthase-pair domain divalent metal cation transport mediator (CNNM)-transient receptor potential ion channel subfamily M member 7 (TRPM7) complex by ADP-ribosylation factor-like GTPase 15 (ARL15) and phosphatases of regenerating liver (PRLs).
CNNM on their own mediate cation efflux. PRL binding to CNNM inhibits cation efflux via CNNMs and activates TRPM7 influx. ARL15 binding to CNNM inhibits both TRPM7 and CNNM transport activity.
Tables
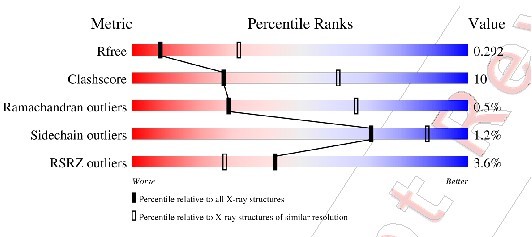
Statistics of data collection and refinement.
| PDB code | 8F6D |
|---|---|
| Data collection | |
| X-ray source | APS 24ID-E |
| Wavelength (Å) | 0.9792 |
| Space group | P1 |
| Cell dimensions | |
| a, b, c (Å) | 66.02, 73.32, 79.33 |
| α, β, γ (°) | 94.4, 95.5, 115.3 |
| Resolution (Å) | 50–3.20 (3.26–3.20)* |
| Rmerge | 0.078 (0.782) |
| I/σI | 10.4 (0.6) |
| Completeness (%) | 94.2 (93.1) |
| Redundancy | 3.7 (3.8) |
| CC1/2 | 0.998 (0.624) |
| Refinement | |
| Resolution (Å) | 29.5–3.20 |
| No. reflections | 20526 |
| Rwork/Rfree | 0.264/0.292 |
| No. atoms | |
| Protein | 7791 |
| B-factors | |
| Protein | 115.3 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.79 |
| Ramachandran statistics (%) | |
| Most favored regions | 97.98 |
| Additional allowed regions | 1.56 |
| Disallowed regions | 0.46 |
-
*
Highest resolution shell is shown in parentheses.