Modulation of sleep by trafficking of lipids through the Drosophila blood-brain barrier
Figures
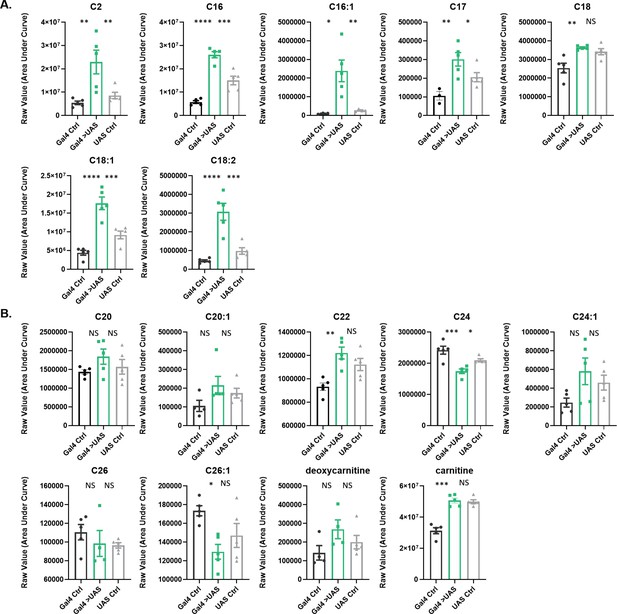
Acylcarnitine levels are increased in Repo>Shi1 fly heads.
(A) Short and long chain length or (B) very long chain length acylcarnitines from Repo>Shi1 fly heads and parental controls. The raw signal from LC/MS is plotted, n=3–5 samples, of 200 fly heads each. One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 1—source data 1
Acylcarnitine measurements in Repo>Shi1 and control fly heads.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig1-data1-v3.xlsx
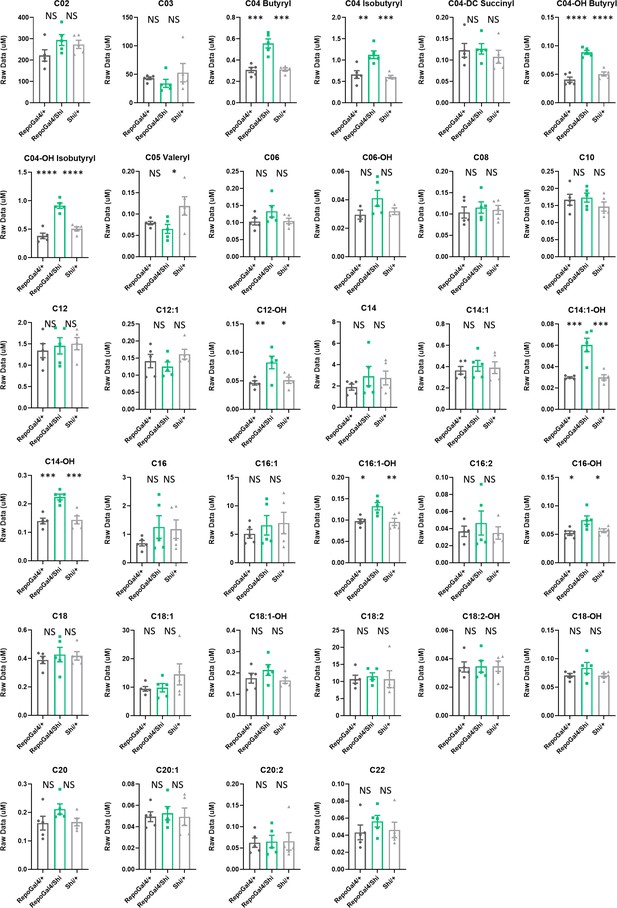
Levels of some acylcarnitines are increased in Repo>Shi1 fly bodies.
Short, long chain and very long chain length acylcarnitines from Repo-GAL4>UAS-Shi1 fly bodies and parental controls. The raw signal from LC-MS is plotted, n=5 samples of 50 fly bodies each. One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 1—figure supplement 1—source data 1
Acylcarnitine measurements in Repo>Shi1 and control fly bodies.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig1-figsupp1-data1-v3.xlsx
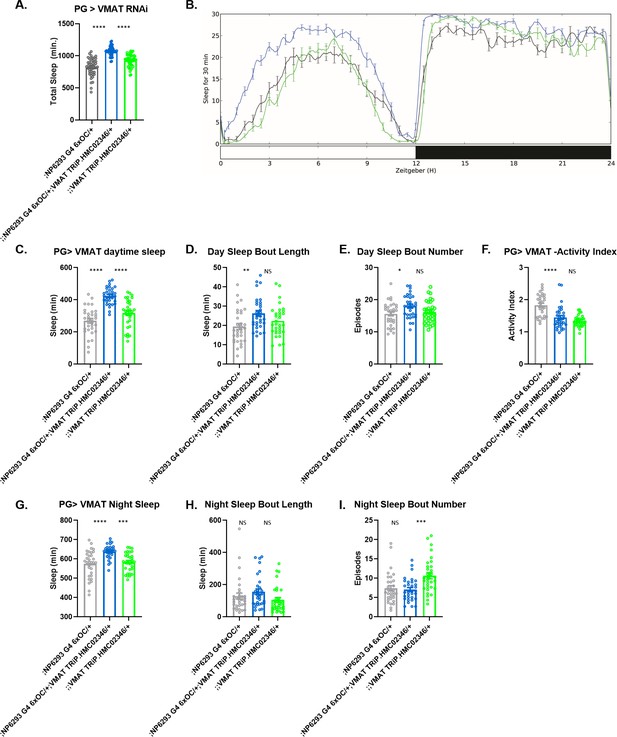
Change in sleep with knockdown of specific barrier-enriched genes in barrier glia.
Total sleep (A and B) and sleep architecture (C–I) in female flies with knockdown of VMAT, UAS-HMC02346 driven by (perineurial glia [PG]) NP6293-GAL4. Mean sleep, sleep bout length and bout number are shown for daytime sleep (C–E) and nighttime sleep (G–I). Activity index, which depicts activity per waking minute, was not different from both controls (F). n=45–48 per genotype. One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 2—source data 1
Sleep data of flies with VMAT knockdown in perineurial glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig2-data1-v3.xlsx
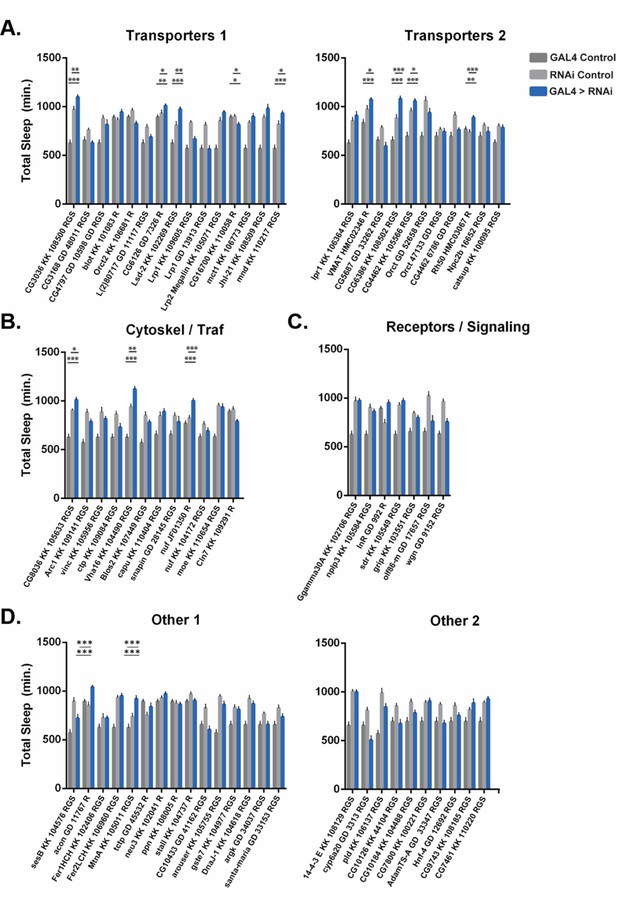
Pan-glial RNAi screen of candidate genes enriched in the barrier glia.
Total sleep in female flies with RNAi knockdown of listed genes (KK and GD are VDRC collections, TRiP lines are from Bloomington Stock Center): (A) Transporters, (B) Cytoskeleton/Trafficking factors, (C) Receptors/Signaling pathway factors, and (D) other genes by either Repo-GAL4 (labeled R) or UAS-Dicer; RepoGeneSwitch on RU+food (labeled RGS). n=9–16 flies per genotype, median = 16. One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01, ***p<0.001. Error bars represent standard error of the mean (SEM). Significance values only marked for genes in which experimental flies were different from both parental controls. Certain experiments were performed simultaneously and therefore share a GAL4 control, re-plotted per each gene.
-
Figure 2—figure supplement 1—source data 1
Sleep data from the pan-glial RNAi screen of candidate genes enriched in the barrier glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig2-figsupp1-data1-v3.xlsx
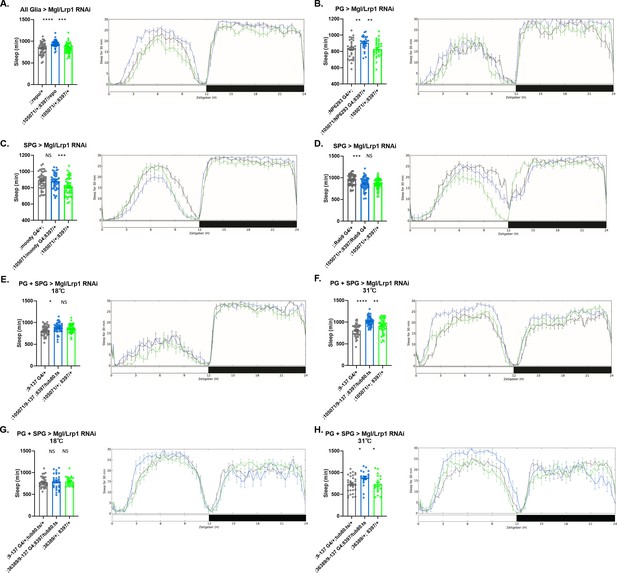
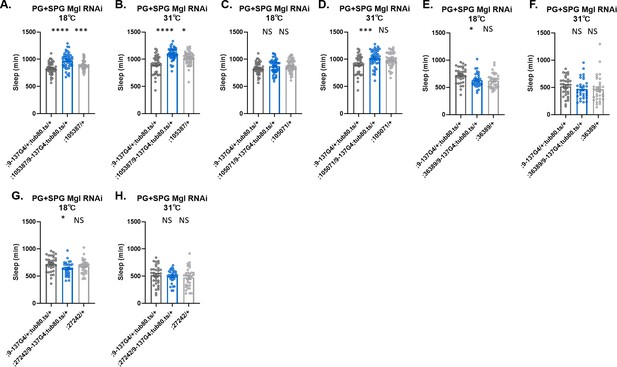
Sleep amount changes with knockdown of Lrp1 and Megalin genes in all glia or barrier glia.
Total sleep in female flies with knockdown of (A) Lrp1 (8397 GD) and Megalin (105071 KK) RNAi driven by Repo-GAL4. n=45–48 per genotype; (B) Lrp1 (8397 GD) and Megalin (105071 KK) RNAi driven by NP6293-GAL4 (perineurial glia [PG]). n=45–48 per genotype; (C) Lrp1 (8397 GD) and Megalin (105071 KK) RNAi driven by moody-GAL4 (subperineurial glia [SPG]) n=45–48 per genotype; (D) Lrp1 (8397 GD) and Megalin (105071 KK) RNAi expressed by Rab9-GAL4 (SPG), n=45–48 per genotype; (E) Lrp1 (8397 GD) and Megalin (105071 KK) RNAi driven by 9–137-GAL4 (PG and SPG) with tub-Gal80ts at the permissive temperature of 18°C, n=45–48 per genotype (F) Lrp1 (8397 GD) and Megalin (105071 KK) RNAi driven by 9–137-GAL4 (SPG and PG) with tub-Gal80ts at the restrictive temperature of 31°C, n=45–48 per genotype. (G) Lrp1 (8397 GD) and Megalin (36389 GD) RNAi driven by 9–137-GAL4 (PG and SPG) with tub-Gal80ts at the permissive temperature of 18°C, n=45–48 per genotype (H) Lrp1 (8397 GD) and Megalin (36389 GD) RNAi driven by 9–137-GAL4 (SPG and PG) with tub-Gal80ts at the restrictive temperature of 31°C, n=45–48 per genotype. The same flies were assayed at permissive and restrictive temperatures. Comparisons were with one-way ANOVA, with Holk-Sidak post hoc. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 3—source data 1
The sleep data of flies with simultaneous knockdown of Lrp1 and Megalin in all glia, perineurial glia , subperineurial glia or in perineurial glia plus subperineurial glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig3-data1-v3.xlsx
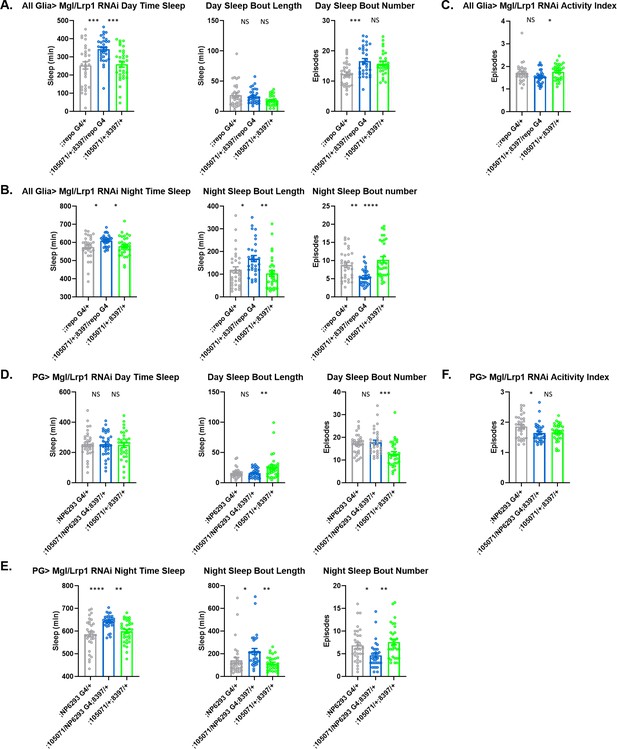
Sleep/activity characteristics with knock down of Lrp in all glia or in perineurial glia (PG).
(A) Mean daytime sleep, bout number and bout length; (B) mean nighttime sleep, bout length and bout number; (C) activity index with knockdown of Megalin (105071 KK) and Lrp1 (8397 GD) in all glia with Repo-GAL4. (D) Mean daytime sleep, bout number and bout length; (E) mean nighttime sleep, bout length and bout number; (F) activity index with knockdown of Megalin (105071 KK) and Lrp1 (8397 GD) in PG with NP6293-GAL4. n=45–48 per genotype. One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 3—figure supplement 1—source data 1
The sleep architecture data of flies with simultaneous knockdown of Lrp1 and Megalin in all glia and perineurial glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig3-figsupp1-data1-v3.xlsx
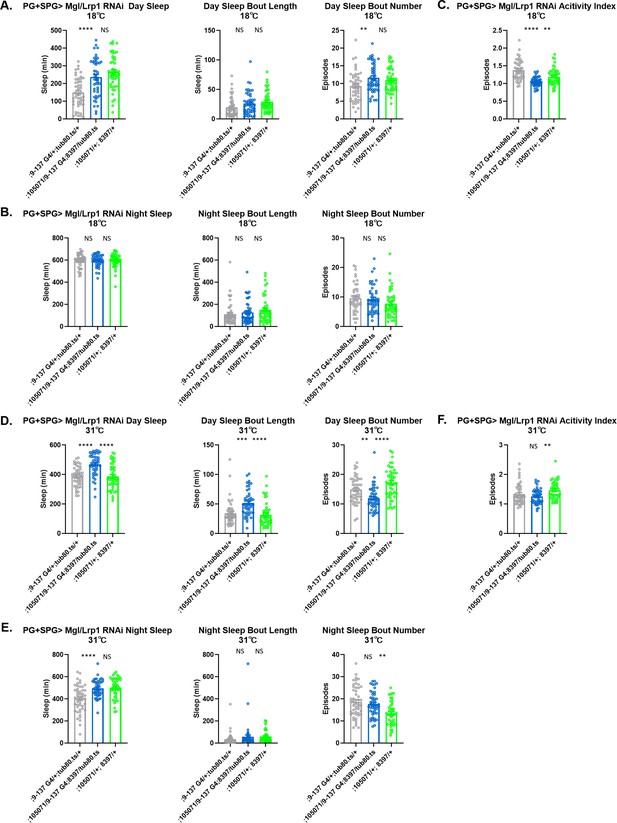
Sleep/activity characteristics with knockdown of Lrp in perineurial glia (PG) and subperineurial glia (SPG).
(A) Mean daytime sleep, bout number and bout length; (B) mean nighttime sleep, bout length and bout number; (C) activity index with knockdown of Megalin (105071 KK) and Lrp1 (8397 GD) in PG and SPG driven by 9–137-GAL4 at 18°C (permissive). (D) Mean daytime sleep, bout number and bout length; (E) mean nighttime sleep, bout length and bout number; (F) activity index with knockdown of Megalin (105071 KK) and Lrp1 (8397 GD) in PG and SPG with 9–137-GAL4 coupled with tub-Gal80ts at 31°C (restrictive). n=45–48 per genotype. One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 3—figure supplement 2—source data 1
The sleep architecture data of flies with simultaneous knockdown of Lrp1 and Megalin in perineurial glia and subperineurial glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig3-figsupp2-data1-v3.xlsx
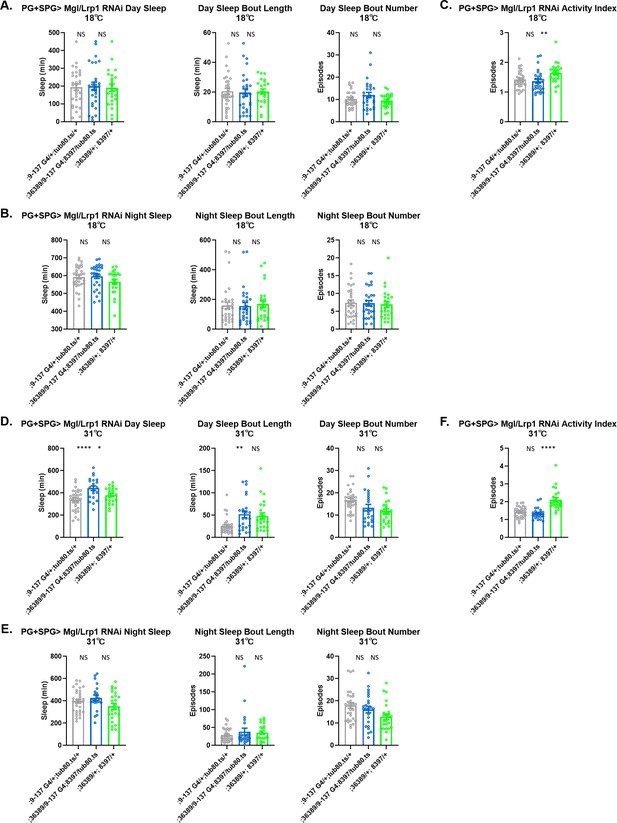
Sleep/activity characteristics with knockdown of Lrp in perineurial glia (PG) and subperineurial glia (SPG).
(A) Mean daytime sleep bout number and bout length; (B) mean nighttime sleep, bout length, bout number; and (C) activity index with knockdown of Megalin (36389 GD) and Lrp1 (8397 GD) in PG and SPG with 9–137-GAL4 at 18°C (permissive temperature). (D) Mean daytime sleep bout number and bout length; (E) mean nighttime sleep, bout length and bout number; (F) activity index with knockdown of Megalin (36389 GD) and Lrp1 (8397 GD) in PG and SPG with 9–137-GAL4 coupled with tub-Gal80ts at 31°C (restrictive). n=45–48 per genotype. One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 3—figure supplement 3—source data 1
The sleep architecture data of flies with simultaneous knockdown of Lrp1(8397 GD) and Megalin (36389 GD) in barrier glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig3-figsupp3-data1-v3.xlsx
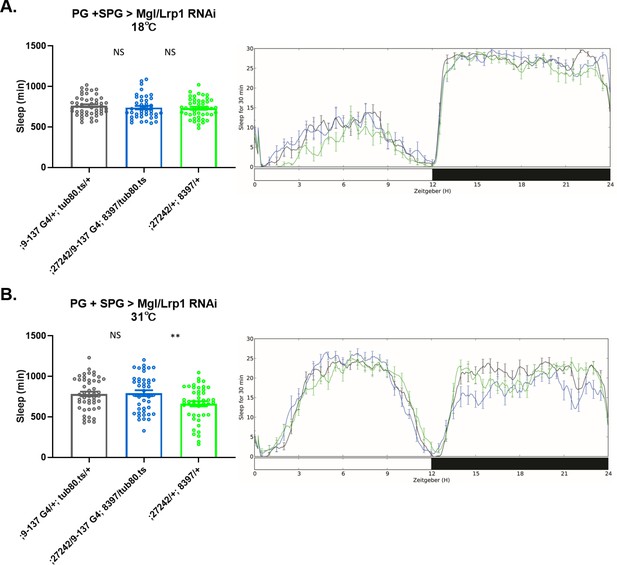
Sleep amount changes with knockdown of Lrp genes in barrier glia.
(A) Lrp1 (8397 GD) and Megalin (27242 GD) RNAi driven by 9–137-GAL4 (perineurial glia [PG] and subperineurial glia [SPG]) with tub-Gal80ts at the permissive temperature of 18°C, n=45–48 per genotype. (B) Lrp1 (8397 GD) and Megalin (27242 GD) RNAi driven by 9–137-GAL4 (SPG and PG) with tub-Gal80ts at the restrictive temperature of 31°C, n=45–48 per genotype. One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01, ****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 3—figure supplement 4—source data 1
The sleep data of flies with simultaneous knockdown of Lrp1(8397 GD) and Megalin (27242 GD) in barrier glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig3-figsupp4-data1-v3.xlsx
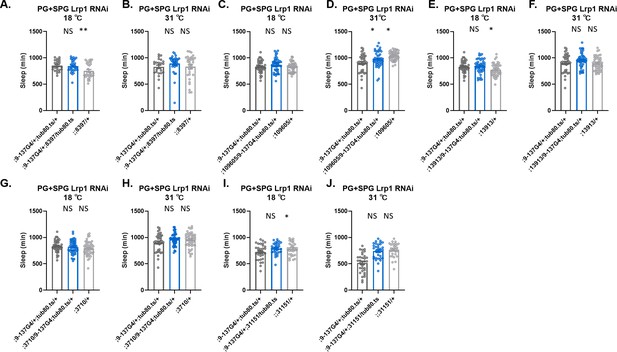
Sleep amount is largely intact with knockdown of single Lrp1 genes in surface glia.
Total sleep in female flies with knockdown of (A and B) Lrp1 (8397 GD), (C and D) Lrp1(109605 KK), (E and F) Lrp1 (13913 KK), (G and H) Lrp1 (3710 GD), (I and J) Lrp1 (31151) driven by (perineurial glia [PG]+subperineurial glia [SPG]) driver 9–137-GAL4 with tub-Gal80ts, at 18°C (permissive) and at 31°C (restrictive). n=32 per genotype. One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01. Error bars represent standard error of the mean (SEM).
-
Figure 3—figure supplement 5—source data 1
The sleep data of flies with single knockdown of Lrp1 in barrier glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig3-figsupp5-data1-v3.xlsx
Sleep amount is largely intact with knockdown of single Megalin genes in surface glia.
Total sleep in female flies with knockdown of (A) Megalin (105387 KK), (C) Megalin (1105071 KK), (E) Megalin (36389 GD), (G) Megalin (27242 GD) at 18°C (permissive) and (B) Megalin (105387 KK), (D) Megalin (1105071 KK), (F) Megalin (36389 GD), (J) Megalin (27242 GD) at 31°C (restrictive). One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 3—figure supplement 6—source data 1
The sleep data of flies with single knockdown of Megalin in barrier glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig3-figsupp6-data1-v3.xlsx
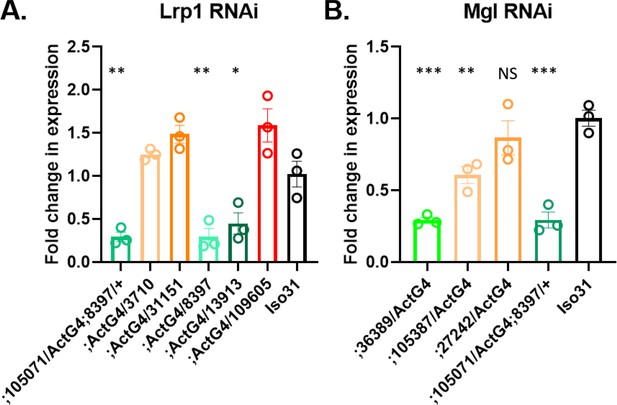
Effect of RNAi constructs on the expression of Lrp1 and Megalin gene.
(A) Lrp1 expression in RNAi flies with an Act-GAL4 driver and in control iso31. (B) Megalin gene expression in RNAi flies with Act-GAL4 driver and in control iso31. Mann-Whitney T test was performed. *p<0.05. Error bars represent standard error of the mean (SEM).
-
Figure 3—figure supplement 7—source data 1
The expression of Lrp1 and Megalin genes in different RNAi fly lines carrying an Act-Gal4 driver.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig3-figsupp7-data1-v3.xlsx
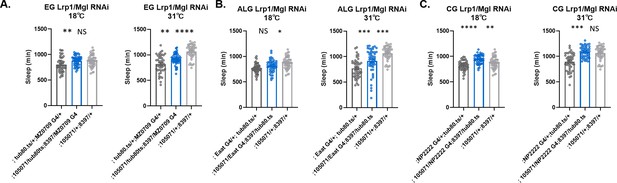
Sleep does not change with simultaneous knockdown of Lrp1 and Megalin genes in ensheathing, astrocyte-like glia, or cortex glia.
Total sleep in female flies with simultaneous knockdown of (A) Lrp1 (8397 GD) and Megalin (105071 KK) in ensheathing glia at 18°C (permissive) and at 31°C (restrictive). (B) Lrp1 (8397 GD) and Megalin (105071 KK) in astrocyte-like glia at 18°C (permissive) and at 31°C (restrictive). (C) Lrp1 (8397 GD) and Megalin (105071 KK) in cortex glia at 18°C (permissive) and at 31°C (restrictive). One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars represent standard error of the mean (SEM). EG, ensheathing glia; CG, cortex glia, ALG, astrocyte-like glia.
-
Figure 3—figure supplement 8—source data 1
The sleep data of flies with simultaneous knockdown of Lrp1 and Megalin in ensheathing, astrocyte-like glia or cortex glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig3-figsupp8-data1-v3.xlsx
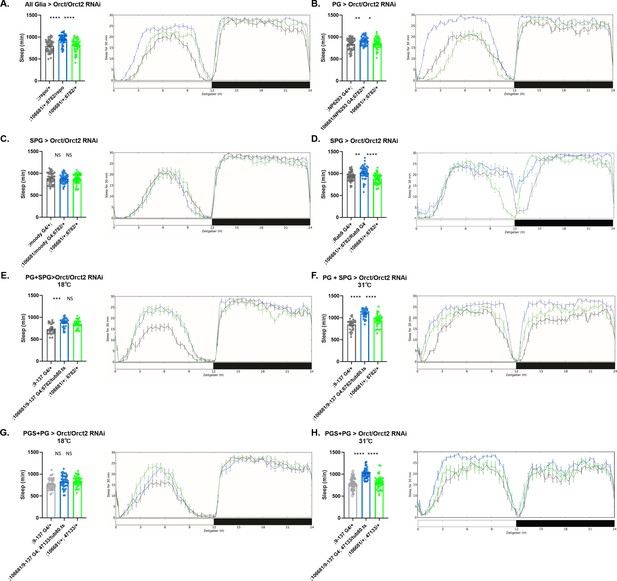
Sleep amount changes with knockdown of Orct genes in all glia or barrier glia.
Total sleep in female flies with knockdown of (A) Orct (6782 GD) and Orct2 (106681 KK) RNAi driven by Repo-GAL4. n=45–48 per genotype; (B) Orct (6782 GD) and Orct2 (106681 KK) RNAi driven by NP6293-GAL4 (perineurial glia [PG]). n=45–48 per genotype; (C) Orct (6782 GD) and Orct2 (106681 KK) RNAi driven by moody-GAL4 (subperineurial glia [SPG]) n=45–48 per genotype; (D) Orct (6782 GD) and Orct2 (106681 KK) RNAi expressed by Rab9-GAL4 (SPG), n=45–48 per genotype; (E) Orct (6782 GD) and Orct2 (106681 KK) RNAi driven by 9–137-GAL4 (PG and SPG) with tub-Gal80ts at the permissive temperature of 18°C, n=45–48 per genotype; (F) Orct (6782 GD) and Orct2 (106681 KK) RNAi driven by 9–137-GAL4 (SPG and PG) with tub-Gal80ts at the restrictive temperature of 31°C, n=45–48 per genotype; (G) Orct (47133 GD) and Orct2 (106681 KK) RNAi driven by 9–137-GAL4 (PG and SPG) with tub-Gal80ts at the permissive temperature of 18°C, n=45–48 per genotype; (H) Orct (47133 GD) and Orct2 (106681 KK) RNAi driven by 9–137-GAL4 (SPG and PG) with tub-Gal80ts at the restrictive temperature of 31°C, n=45–48 per genotype. Comparisons were with one-way ANOVA, followed by Holk-Sidak post hoc. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 4—source data 1
The sleep data of fiies that had simultaneous knock down of Orct and Orct2 in all glia or barrier glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig4-data1-v3.xlsx
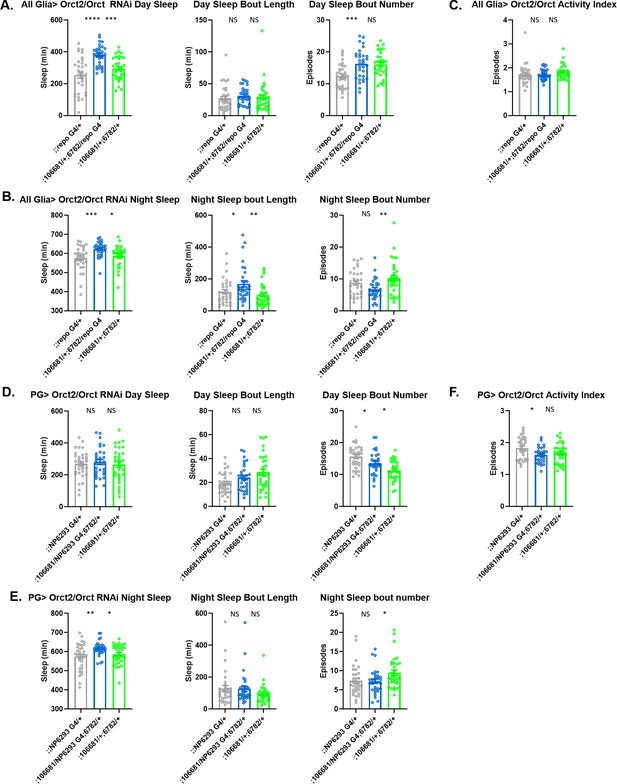
Sleep/activity characteristics with knockdown of Orct in all glia or in perineurial glia (PG).
(A) Mean daytime sleep, bout length and bout number, (B) mean nighttime sleep, bout length and bout number, and (C) activity index with knockdown of Orct2 (106681 KK) and Orct (6782 GD) in all glia with Repo-GAL4; (D) mean daytime sleep, bout length and sleep bout number, (E) mean nighttime sleep, bout length and bout number, and (F) activity index with knockdown of Orct2 (106681 KK) and Orct (6782 GD) in PG with NP6293-GAL4. n=45–48 per genotype. One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 4—figure supplement 1—source data 1
The sleep/activity architecture data of flies that had simultaneous knock down of Orct and Orct2 in all glia or perineurial glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig4-figsupp1-data1-v3.xlsx
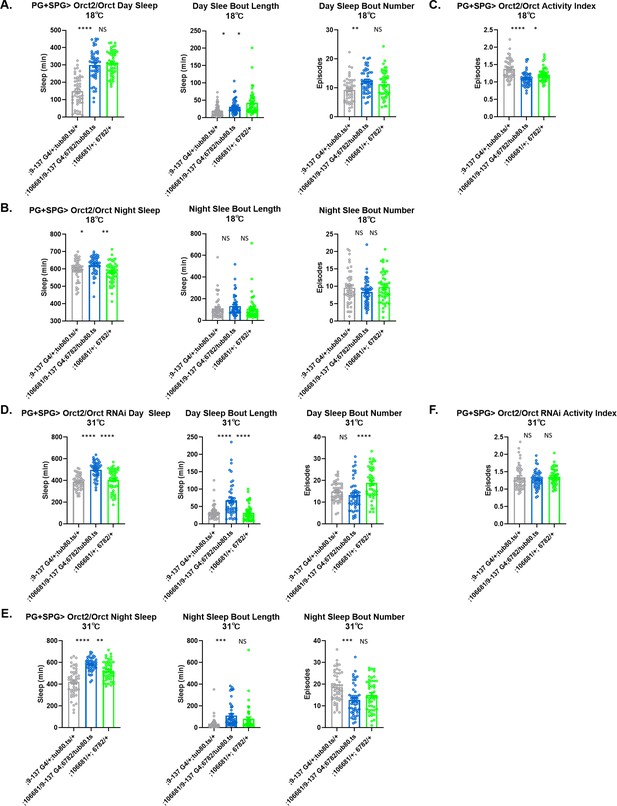
Sleep/activity characteristics with knock down of Orct in perineurial glia (PG) and subperineurial glia (SPG).
(A) Mean daytime sleep, bout length and bout number, (B) mean nighttime sleep, bout length and bout number, and (C) activity index with knockdown of Orct2 (106681 KK) and Orct (6782 GD) in PG and SPG with 9–137-GAL4 at 18°C (permissive); (D) mean daytime sleep, bout number and bout length, (E) mean nighttime sleep, bout length and bout number, and (F) activity index with knockdown of Orct2 (106681 KK) and Orct (6782 GD) in PG and SPG with 9–137-GAL4 coupled with tub-Gal80ts at 31°C (restrictive). n=45–48 per genotype. One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 4—figure supplement 2—source data 1
Sleep and sleep architecture data produced by knockdown Orct (6782 GD) and Orct2 (106681 KK) in barrier glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig4-figsupp2-data1-v3.xlsx
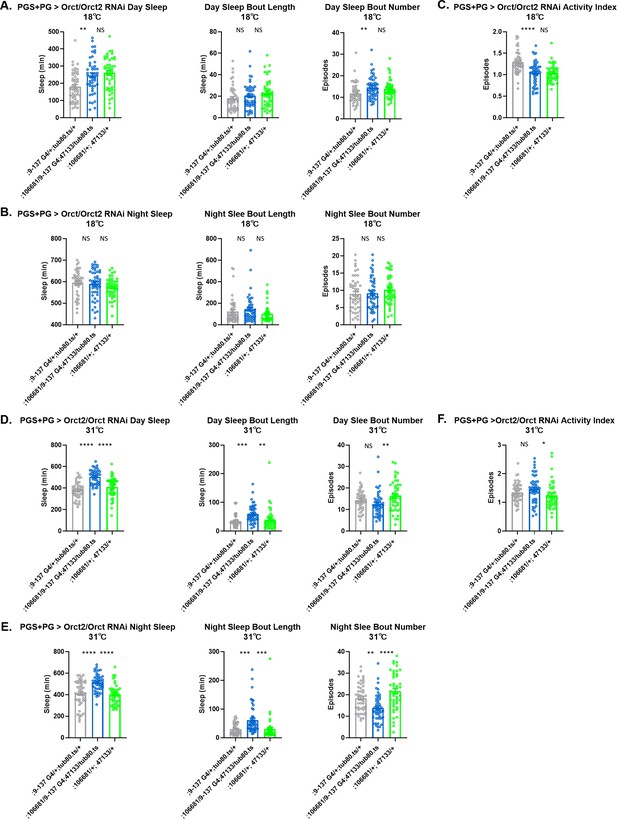
Sleep/activity characteristics with knockdown of Orct in perineurial glia (PG) and subperineurial glia (SPG).
(A) Mean daytime sleep, sleep bout length and sleep bout number, (B) mean nighttime sleep, bout length and bout number, and (C) activity index with knockdown of Orct2 (106681 KK) and Orct (47133 GD) in PG and SPG with 9–137-GAL4 coupled with tub-Gal80ts at 18°C (permissive); (D) mean daytime sleep, bout length and bout number, (E) mean nighttime sleep, sleep bout length and bout number, and (F) activity index with knockdown of Orct2 (106681 KK) and Orct (47133 GD) in PG and SPG with 9–137-GAL4 coupled with tub-Gal80ts at 31°C (restrictive). n=45–48 per genotype. One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 4—figure supplement 3—source data 1
Sleep and sleep architecture data produced by knockdown of Orct (47133 GD) and Orct2 (106681 KK) in barrier glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig4-figsupp3-data1-v3.xlsx
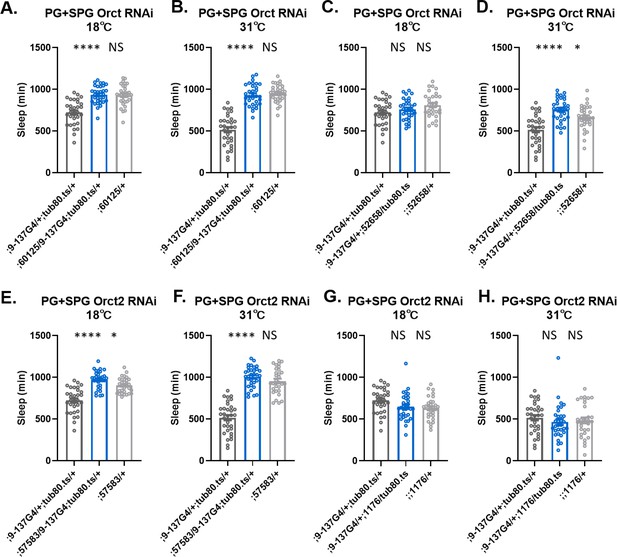
Sleep amount is largely intact with knockdown of single Orct genes in surface glia.
Total sleep in female flies with knockdown of (A) Orct (60125), (C) Orct (52658 GD), (E) Orct2 (57583), (G) Orct2 (1176 GD) at 18°C (permissive) and (B) Orct (60125), (D) Orct (52658 GD), (F) Orct2 (57583), (H) Orct2 (1176 GD) at 31°C (restrictive). One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01, ***p<0.001,****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 4—figure supplement 4—source data 1
Sleep data of flies with single knockdown of Orct or Orct2 in barrier glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig4-figsupp4-data1-v3.xlsx
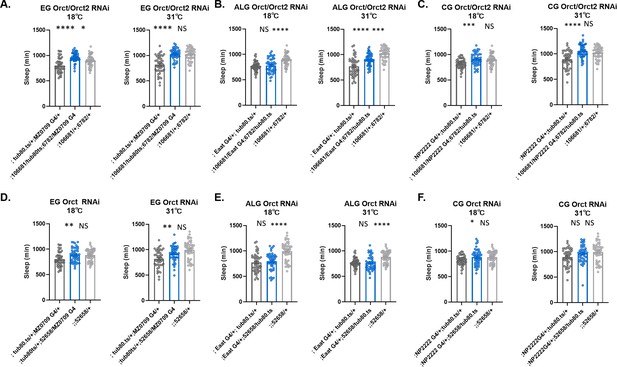
Sleep does not change with simultaneous knockdown of Orct and Orct2 genes in ensheathing, astrocyte-like glia, or cortex glia.
Total sleep in female flies with knockdown of (A) Orct (6782 GD) and Orct2 (106681 KK) in ensheathing glia at 18°C (permissive) and at 31°C (restrictive). (B) Orct (6782 GD) and Orct2 (106681 KK) in astrocyte-like glia at 18°C (permissive) and at 31°C (restrictive). (C) Orct (6782 GD) and Orct2 (106681 KK) in cortex glia at 18°C (permissive) and at 31°C (restrictive). (D) Orct (6782 GD) and Orct2 (106681 KK) in ensheathing glia at 18°C (permissive) and at 31°C (restrictive). (E) Orct (52658 GD) in astrocyte-like glia at 18°C (permissive) and at 31°C (restrictive). (F) Orct (52658 GD) in cortex glia at 18°C (permissive) and at 31°C (restrictive). One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars represent standard error of the mean (SEM). EG, ensheathing glia; CG, cortex glia; ALG, astrocyte-like glia.
-
Figure 4—figure supplement 5—source data 1
Sleep data of flies with simultaneous knock down of Orct and Orct2 in ensheathing, astrocyte-like glia or cortex glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig4-figsupp5-data1-v3.xlsx
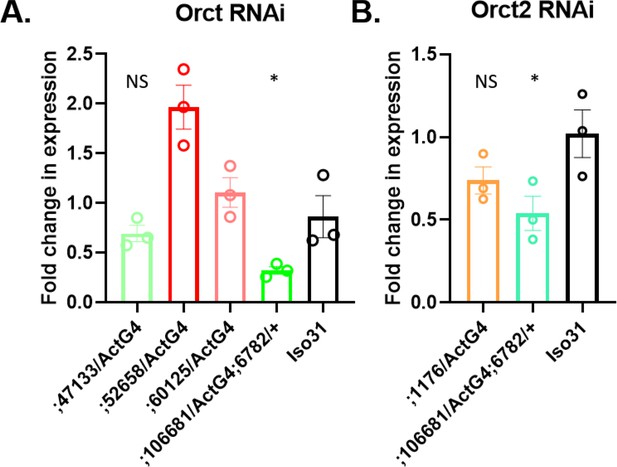
Expression of Orct and Orct2 genes in RNAi fly lines.
(A) Orct gene expression in RNAi fly lines using an Act-GAL4 driver and in control iso31. (B) Orct2 gene expression in RNAi fly lines with an Act-GAL4 driver and in control iso31. Mann-Whitney T test was performed. *p<0.05. Error bars represent standard error of the mean (SEM).
-
Figure 4—figure supplement 6—source data 1
The expression of Orct and Orct2 RNA in fly lines where Orct or Orct2 RNAi were driven by Act-Gal4 driver.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig4-figsupp6-data1-v3.xlsx
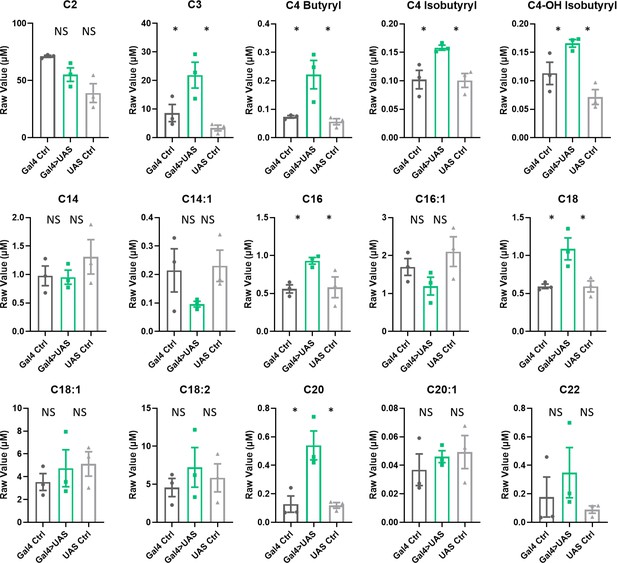
Acylcarnitine levels are increased in Repo>Lrp1+Mgl RNAi fly heads.
Short, medium, and long chain length acylcarnitines from Repo-GAL4>Lrp1+Mgl RNAi fly heads and parental controls. The raw signal from LC/MS is plotted, n=3 samples, of 300 fly heads each. One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05, **p<0.01. Error bars represent standard error of the mean (SEM).
-
Figure 5—source data 1
Acylcarnitine measurements in fly heads with simultaneous knockdown of Lrp1 and Megalin in all glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig5-data1-v3.xlsx
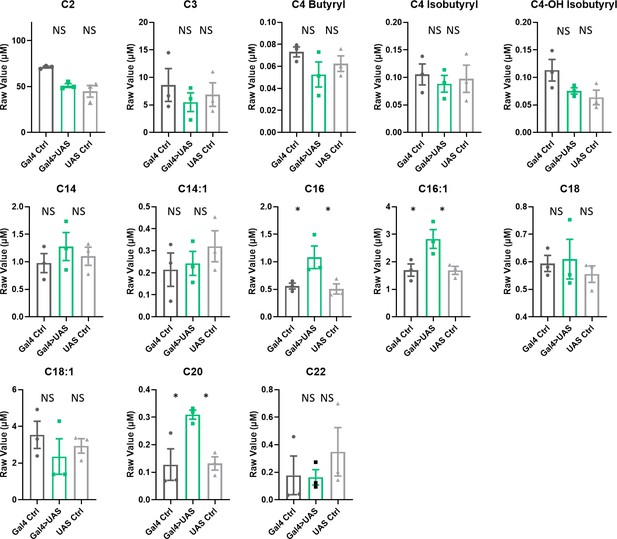
Acylcarnitine levels are increased in Repo>Orct2+Orct RNAi fly heads.
Short, medium, and long chain length acylcarnitines from Repo-GAL4>Orct + Orct2 RNAi fly heads and parental controls. The raw signal from LC/MS is plotted, n=3 samples, of 300 fly heads each. One-way ANOVA, with Holk-Sidak post hoc comparisons. *p<0.05. Error bars represent standard error of the mean (SEM).
-
Figure 6—source data 1
Acylcarnitine measurements in fly heads with simultaneous knockdown of Orct and Orct2 in all glia.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig6-data1-v3.xlsx
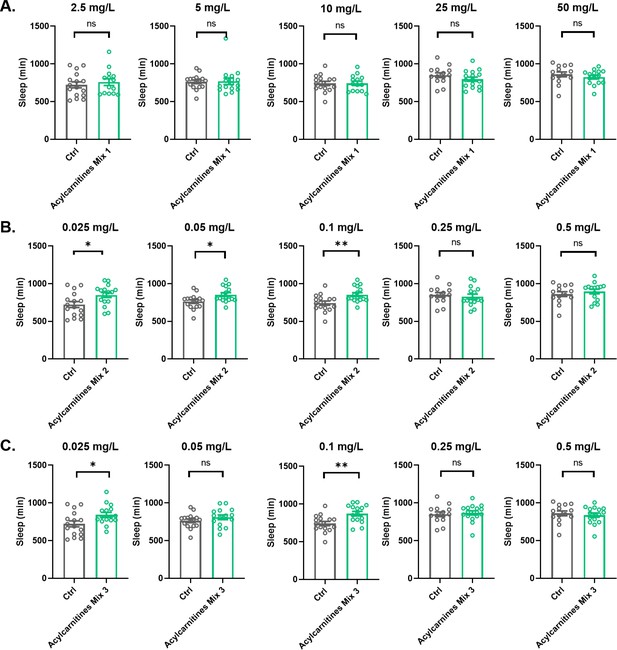
Effects of acylcarnitine Mix 1, 2, or 3 on sleep.
Total sleep was evaluated in female iso31 flies fed with mixes of short or long chain length acylcarnitines or control food. (A) Acylcarnitine Mix 1 at 2.5 mg/L, 5 mg/L, 10 mg/L, 25 mg/L, and 50 mg/L concentrations. (B) Acylcarnitine Mix 2 at 0.025 mg/L, 0.05 mg/L, 0.10 mg/L, 0.25 mg/L, and 0.5 mg/L concentrations. (C) Acylcarnitine Mix 3 at 0.025 mg/L, 0.05 mg/L, 0.10 mg/L, 0.25 mg/L, and 0.5 mg/L concentrations. N=16 per condition. Mann-Whitney T test was performed. *p<0.05, **p<0.01. Error bars represent standard error of the mean (SEM).
-
Figure 7—source data 1
Sleep data of flies fed food containing acylcarnitines Mix 1, 2 or 3 or control solvent.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig7-data1-v3.xlsx
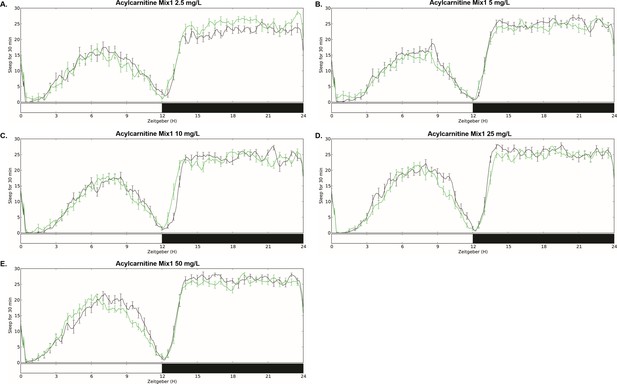
Sleep amount with acylcarnitine Mix 1 feeding.
Daily baseline sleep amount (3 day mean) in female iso31 flies fed with mixes of short chain length acylcarnitine Mix 1 at (A) 2.5 mg/L, (B) 5 mg/L, (C) 10 mg/L, (D) 25 mg/L, or 50 mg/L concentrations (green) or control food (black).
-
Figure 7—figure supplement 1—source data 1
Sleep data of flies fed food containing acylcarnitine Mix 1 or control solvent.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig7-figsupp1-data1-v3.xlsx
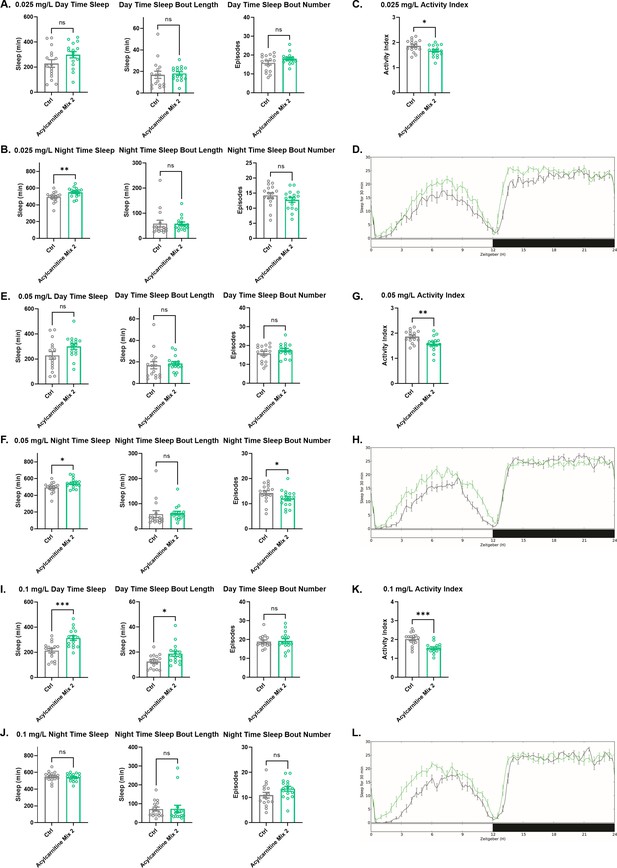
Sleep/activity characteristics with feeding of low concentrations of acylcarnitine Mix 2.
(A) Mean daytime sleep, bout length and bout number, (B) mean nighttime sleep, bout length and bout number, (C) activity index and (D) daily baseline sleep amount (3 day mean) with feeding of 0.025 mg/L acylcarnitine Mix 2; (E) mean daytime sleep, bout length and bout number, (F) mean nighttime sleep, bout length and bout number, (G) activity index and (H) daily baseline sleep amount (3 day mean) with feeding of 0.05 mg/L acylcarnitine Mix 2; (I) mean daytime sleep, bout length bout and number, (J) mean nighttime sleep, bout length and bout number, (K) activity index, and (L) daily baseline sleep amount (3 day mean) with feeding of 0.1 mg/L acylcarnitine Mix 2. n=16 per genotype. Mann-Whitney T test was performed. *p<0.05, **p<0.01, ***p<0.001. Error bars represent standard error of the mean (SEM).
-
Figure 7—figure supplement 2—source data 1
Sleep data of flies fed food containing low concentration of acylcarnitine Mix 2 or control solvent.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig7-figsupp2-data1-v3.xlsx
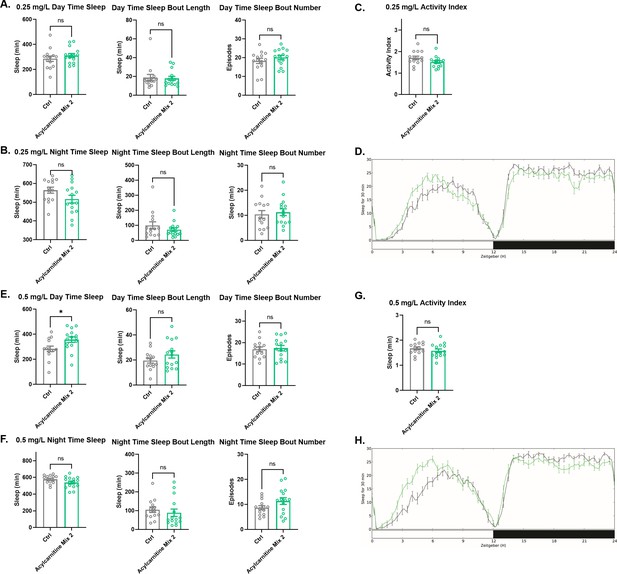
Sleep/activity characteristics with feeding of high concentrations of acylcarnitine Mix 2.
(A) Mean daytime sleep, bout number and bout length, (B) mean nighttime sleep, bout length and bout number, and (C) activity index, (D) daily baseline sleep amount (3 day mean) with feeding of 0. 25 mg/L acylcarnitine Mix 2; (E) mean daytime sleep, bout length and bout number, (F) mean nighttime sleep, bout length and bout number, (G) activity index, and (H) daily baseline sleep amount (3 day mean) with feeding of 0.5 mg/L acylcarnitine Mix 2. n=32 per genotype. Mann-Whitney T test was performed. *p<0.05, **p<0.01, ***p<0.001. Error bars represent standard error of the mean (SEM).
-
Figure 7—figure supplement 3—source data 1
Sleep data of flies fed food containing high concentration of acylcarnitine Mix 2 or control solvent.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig7-figsupp3-data1-v3.xlsx
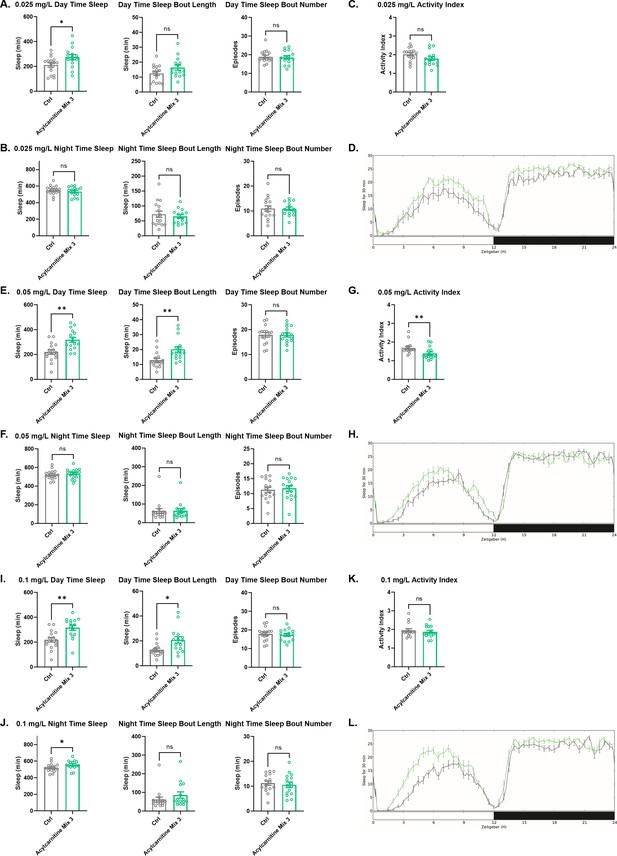
Sleep/activity characteristics with feeding of low concentrations of acylcarnitine Mix 3.
(A) Mean daytime sleep, bout length and bout number, (B) mean nighttime sleep, bout length and bout number, (C) activity index, and (D) daily baseline sleep amount (3 day mean) with feeding of 0.025 mg/L acylcarnitine Mix 3; (E) mean daytime sleep, bout length and bout number, (F) mean nighttime sleep, bout length and bout number, (G) activity index, and (H) daily baseline sleep amount (3 day mean) with feeding of 0.05 mg/L acylcarnitine Mix 3; (I) mean daytime sleep, bout length and bout number, (J) mean nighttime sleep, bout length and bout number, (K) activity index, and (L) daily baseline sleep amount (3 day mean) with feeding of 0.1 mg/L acylcarnitine Mix 3. n=16 per genotype. Mann-Whitney T test was performed. *p<0.05, **p<0.01. Error bars represent standard error of the mean (SEM).
-
Figure 7—figure supplement 4—source data 1
Sleep data of flies fed with food containing low concentration of acylcarnitine Mix 3 or control solvent.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig7-figsupp4-data1-v3.xlsx
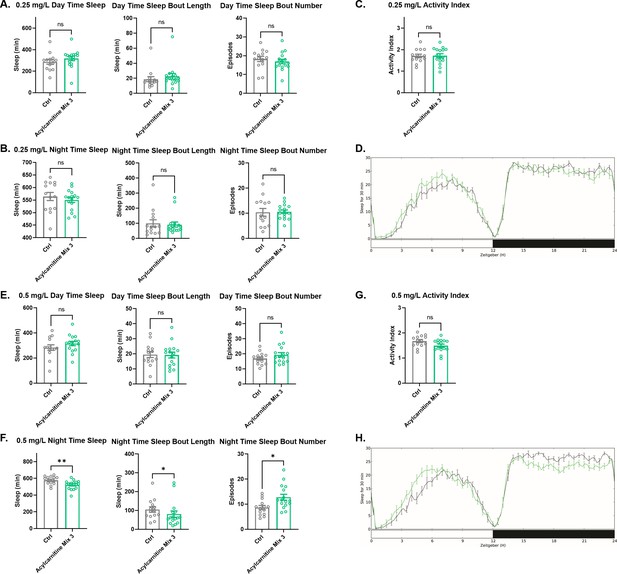
Sleep/activity characteristics with feeding of high concentration of acylcarnitine Mix 3.
(A) Mean daytime sleep, bout length and bout number, (B) mean nighttime sleep, bout length and bout number, (C) activity index, and (D) daily baseline sleep amount (3 day mean) with feeding of 0. 25 mg/L acylcarnitine Mix 3; (E) mean daytime sleep, bout length and bout number, (F) mean nighttime sleep, bout length and bout number, (G) activity index, and (H) daily baseline sleep amount (3 day mean) with feeding of 0.5 mg/L acylcarnitine Mix 3. n=16 per genotype. Mann-Whitney T test was performed. *p<0.05, **p<0.01. Error bars represent standard error of the mean (SEM).
-
Figure 7—figure supplement 5—source data 1
Sleep data of flies fed with food containing high concentration of acylcarnitine Mix 3 or control solvent.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig7-figsupp5-data1-v3.xlsx
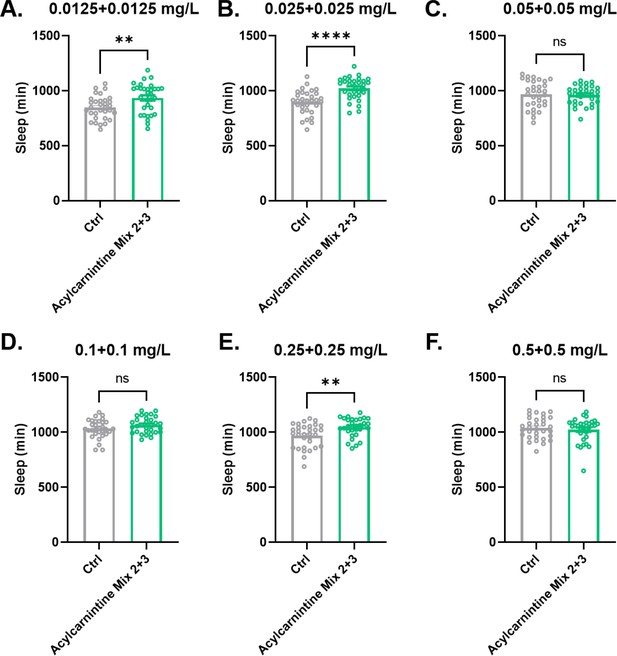
Effects of acylcarnitine Mix 2 and 3 on sleep.
Total sleep was evaluated in female iso31flies fed with mixes of long chain length acylcarnitines or control food. Each of acylcarnitine Mix 2 and 3 was added at a concentration of 0.0125 mg/L (A), 0.025 mg/L (B), 0.05 mg/L (C), 0.10 mg/L (D), 0.25 mg/L (E), or 0.5 mg/L (F). n=32 per condition. Mann-Whitney T test was performed. *p<0.05, **p<0.01, ****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 8—source data 1
Sleep data of flies fed with food containing acylcarnitines Mix 2 and 3 or control solvent.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig8-data1-v3.xlsx
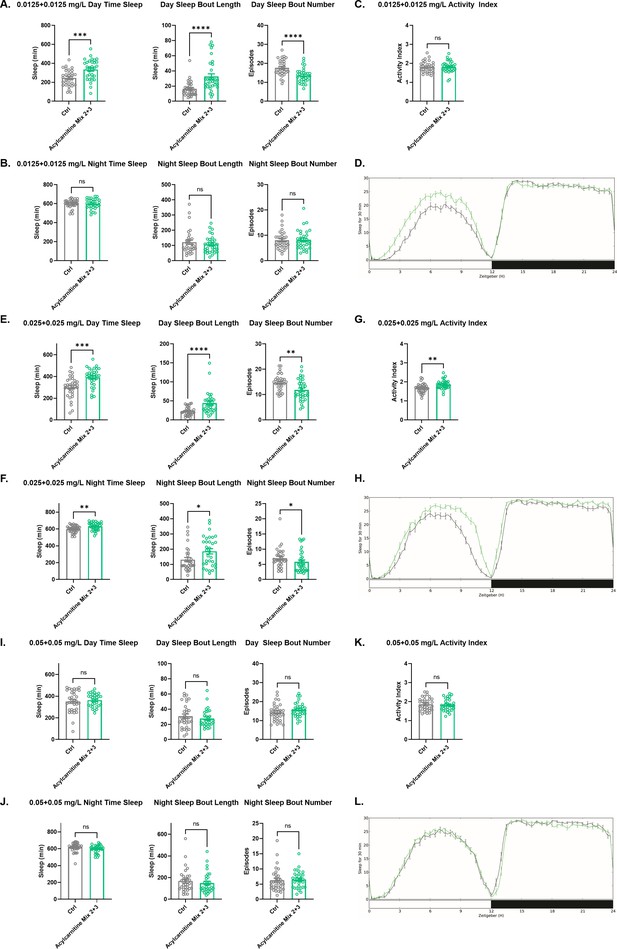
Sleep/activity characteristics with feeding of low concentrations of acylcarnitine Mix 2 and Mix 3.
(A) Mean daytime sleep, bout length and bout number, (B) mean nighttime sleep, bout length and bout number, (C) activity index, and (D) daily baseline sleep amount (3 day mean) with feeding of acylcarnitine Mix 2 and Mix 3 at 0.0125 mg/L each; (E) mean daytime sleep, bout length and bout number, (F) mean nighttime sleep, bout length and bout number, (G) activity index, and (H) daily baseline sleep amount (3 day mean) with 0.025 mg/L of each of acylcarnitine Mix 2 and Mix 3; (I) mean daytime sleep, bout length and bout number, (J) mean nighttime sleep, bout length and bout number, (K) activity index, and (L) daily baseline sleep amount (3 day mean) with 0.05 mg/L of each of acylcarnitine Mix 2 and Mix 3. n=32 per genotype. Mann-Whitney T test was performed. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars represent standard error of the mean (SEM).
-
Figure 8—figure supplement 1—source data 1
The sleep/activity architecture of flies fed with food containing low concentrations of acylcarnitines Mix 2 and 3 or control solvent.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig8-figsupp1-data1-v3.xlsx
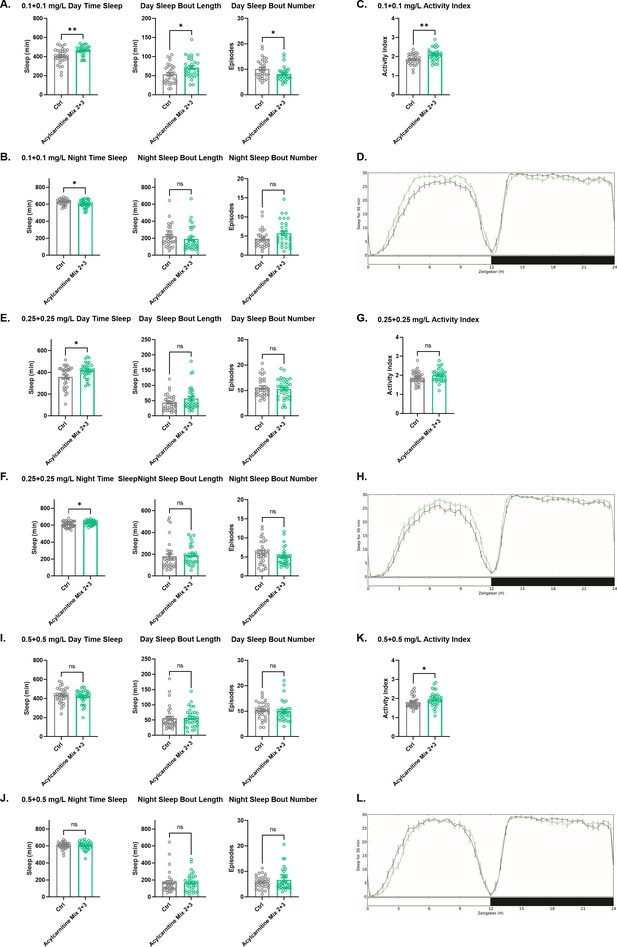
Sleep/activity characteristics with feeding of high concentrations of acylcarnitine Mix 2 and Mix 3.
(A) Mean daytime sleep, bout length and bout number, (B) mean nighttime sleep, bout length and bout number, (C) activity index, and (D) daily baseline sleep amount (3 day mean) with feeding of acylcarnitine Mix 2 and Mix 3, each at 0.1 mg/L; (E) mean daytime sleep, bout length and bout number, (F) mean nighttime sleep, bout length and bout number, (G) activity index, and (H) daily baseline sleep amount (3 day mean) with feeding of 0.25 mg/L of each of acylcarnitine Mix 2 and Mix 3; (I) mean daytime sleep, bout length and bout number, (J) mean nighttime sleep, bout length and bout number, (K) activity index, and (L) daily baseline sleep amount (3 day mean) with 0.5 mg/L of each of acylcarnitine Mix 2 and Mix 3. n=32 per genotype. Mann-Whitney T test was performed. *p<0.05, **p<0.01. Error bars represent standard error of the mean (SEM).
-
Figure 8—figure supplement 2—source data 1
The sleep/activity architecture of flies fed with food containing high concentrations of acylcarnitines Mix 2 and 3 or control solvent.
- https://cdn.elifesciences.org/articles/86336/elife-86336-fig8-figsupp2-data1-v3.xlsx
Tables
Fatty acid acylcarnitine accumulation in Repo>Shi1 fly heads.
Acylcarnitine species in Repo>Shi1 relative to parental controls. Welch’s t-test was performed on the scaled signal for each metabolite, comparing the conditions shown.
| Sub pathway | Biochemical name | |||
|---|---|---|---|---|
| Fatty acid metabolism (acylcarnitine) | Acetylcarnitine (C2) | 4.25 | 2.68 | 1.59 |
| Myristoylcarnitine (C14) | 28.81 | 26.39 | 1.09 | |
| Palmitoylcarnitine (C16) | 4.52 | 1.74 | 2.60 | |
| Palmitoleoycarnitine (C16:1) | 26.50 | 10.35 | 2.56 | |
| Margaroylcarnitine (C17) | 4.05 | 1.47 | 2.76 | |
| Stearoylcarnitine (C18) | 1.43 | 1.06 | 1.34 | |
| Linoleoylcarnitine (C18:2) | 6.77 | 3.14 | 2.15 | |
| Oleoylcarnitine (C18:1) | 4.00 | 1.92 | 2.08 | |
| Arachidoylcarnitine (C20) | 1.28 | 1.17 | 1.09 | |
| Behenoylcarnitine (C22) | 1.31 | 1.09 | 1.20 | |
| Eicosenoylcarnitine (C20:1) | 2.28 | 1.24 | 1.84 | |
| Lignoceroylcarnitine (C24) | 0.72 | 0.83 | 0.87 | |
| Margaroylcarnitine (C17) | 4.05 | 1.47 | 2.76 | |
| Nervonoylcarnitine (C24:1) | 2.36 | 1.26 | 1.87 | |
| Cerotoylcartinine (C26) | 0.85 | 0.97 | 0.87 | |
| Ximenoylcarnitine (C26:1) | 0.75 | 0.88 | 0.85 |
Additional files
-
Supplementary file 1
Metabolomics of Repo>20xShibire fly heads.
All measured metabolites and their respective categories are listed for samples from Repo-GAL4>UAS-20xShibire, and both parental controls. Welch’s t-test was performed on scaled signal for each metabolite, comparing the conditions shown. Green highlighting marks a significant difference (p≤0.05) between the groups, where metabolite ratio is <1.00, while light green is not significant, but close to the threshold (0.05<p<0.10). Red highlighting marks a significant difference (p≤0.05) between groups where metabolite ratio is ≥1.00, and light red is not significant, but close to the threshold (0.05<p<0.10).
- https://cdn.elifesciences.org/articles/86336/elife-86336-supp1-v3.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/86336/elife-86336-mdarchecklist1-v3.docx





