ZMYM2 controls human transposable element transcription through distinct co-regulatory complexes
Figures
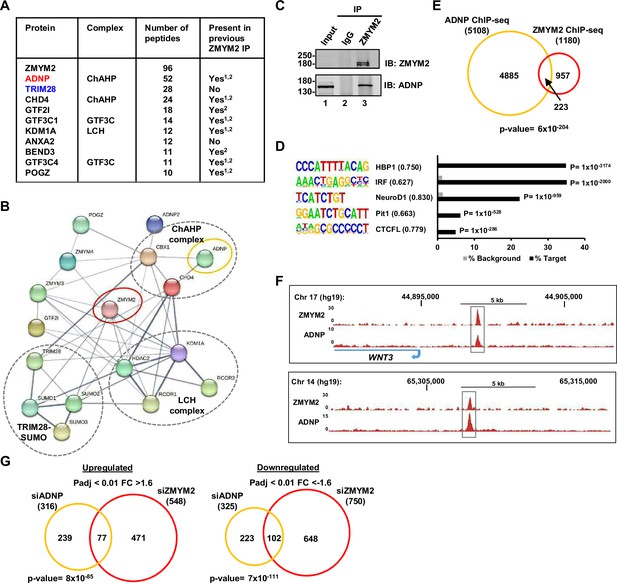
RIME analysis identifies ADNP as a co-regulatory partner of ZMYM2.
(A) Summary table of the ten top scoring interactors for ZMYM2. The average number of peptides across three RIME experiments are shown and whether detected previously in ZMYM2 IP-mass spectrometry experiments is indicated (Yang et al., 2020; Connaughton et al., 2020). Core members of the ChAHP and GTF3C complexes are highlighted. (B) Depiction of interactions between ZMYM2 binding partners found in RIME experiments with known previous molecular interactions found in the STRING database (Jensen et al., 2009). (C) Co-immunoprecipitation analysis of ADNP with ZMYM2. Immunoprecipitation (IP) was performed with ZMYM2 or control IgG antibody from U2OS cells and resulting proteins detected by immunoblotting (IB) with the indicated antibodies. Molecular weight markers (kDa) and 10% input are shown. (D) De novo motif analysis of ADNP binding regions. The top five most significantly enriched motifs are shown, along with motif similarity to the indicated protein in brackets. (E) Venn diagram showing the overlap between ZMYM2 and ADNP binding regions. (F) UCSC genome browser of ChIP-seq data on example genomic loci showing binding of both ZMYM2 and ADNP. (G) Venn diagram showing overlaps in genes upregulated (left) or downregulated (right) following ADNP (orange) or ZMYM2 (red) depletion (fold change >1.6; Padj <0.01). See also Figure 1—figure supplement 1.
-
Figure 1—source data 1
Raw unedited images of Co-immunoprecipitation analysis of ADNP with ZMYM2 (Figure 1C).
Resulting proteins were detected by immunoblotting (IB) with ZMYM2 and ADNP antibodies. The regions used for creating the final figure are boxed. Molecular weight marker sizes (kDa) are shown on the left.
- https://cdn.elifesciences.org/articles/86669/elife-86669-fig1-data1-v1.zip
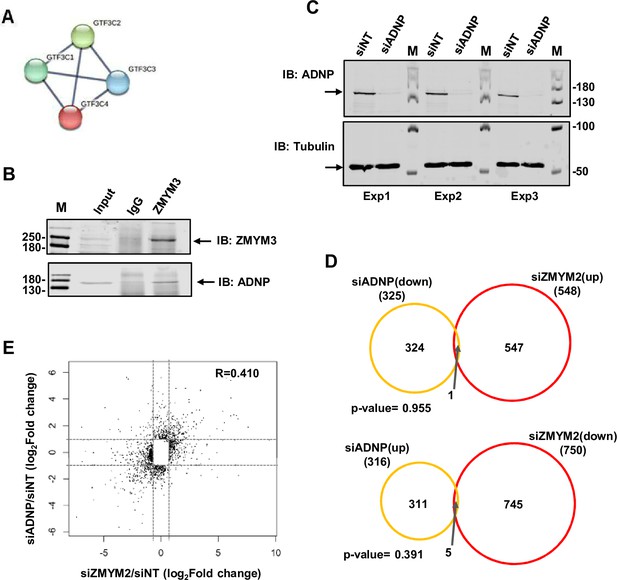
ADNP interactions with ZMYM2.
(A) Depiction of interactions between ZMYM2 binding partners found in RIME experiments with known previous protein interactions in the GTF3C complex found in the STRING database (Jensen et al., 2009). (B) Co-immunoprecipitation analysis of ADNP with ZMYM3. Immunoprecipitation (IP) was performed with ZMYM3 or control IgG antibody from U2OS cells and resulting proteins detected by immunoblotting (IB) with the indicated antibodies. Molecular weight markers (M) and 10% input are shown. (C) Western blot of ADNP expression in U2OS cells following treatment with siADNP or a non-targeting (NT) siRNA (top). Tubulin was used as a loading control (bottom). Molecular weight markers (M) are shown. (D) Venn diagrams showing overlaps in genes showing reciprocal directionality following ADNP (left; orange) or ZMYM2 (right; red) depletion. Genes downregulated with siADNP and upregulated with siZMYM2 (top) or upregulated with siADNP and downregulated with siZMYM2 (bottom) are shown (fold change >1.6; Padj <0.01). (E) Scatterplot of significantly changing genes following ZMYM2 (x-axis) or ADNP2 (y-axis) depletion (fold change >1.6; Padj <0.01).
-
Figure 1—figure supplement 1—source data 1
Raw unedited images of Co-immunoprecipitation analysis of ADNP with ZMYM3 (Figure 1—figure supplement 1B).
Resulting proteins were detected by immunoblotting (IB) with ZMYM3 and TRIM28 antibodies. The regions used for creating the final figure are boxed. Molecular weight marker sizes (kDa) are shown on the left.
- https://cdn.elifesciences.org/articles/86669/elife-86669-fig1-figsupp1-data1-v1.zip
-
Figure 1—figure supplement 1—source data 2
Raw unedited images of Western blot of ADNP expression in U2OS cells following treatment with siADNP or a non-targeting (NT) siRNA (top) (Figure 1—figure supplement 1C).
αTubulin was used as a loading control (bottom). Molecular weight markers (M) are shown. The regions used for creating the final figure are boxed. Molecular weight marker sizes (kDa) are shown on the right.
- https://cdn.elifesciences.org/articles/86669/elife-86669-fig1-figsupp1-data2-v1.zip
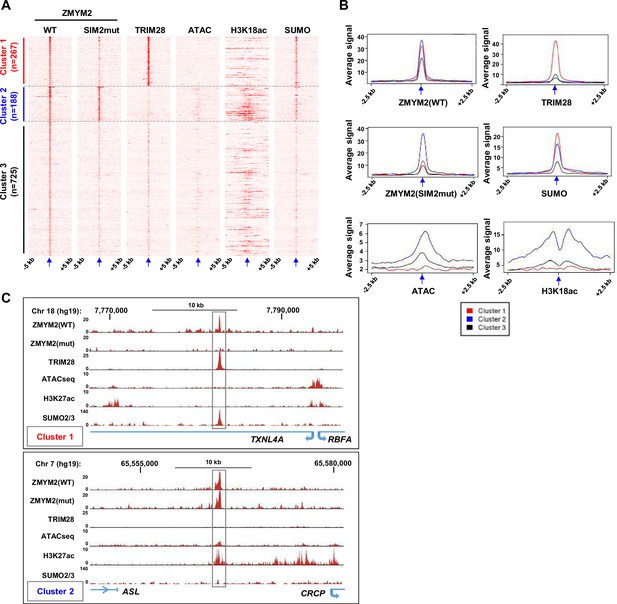
ZMYM2 interactions with molecularly distinct chromatin regions.
(A) Heatmaps showing the signals of the indicated proteins or chromatin marks from ChIP-seq experiments or the ATAC-seq signal in U2OS cells plotted across a 10 kb region surrounding the centres (arrowed) of the wild-type (WT) ZMYM2 binding regions. Clustering of the data produced 3 clusters. (B) Tag density plots of the indicated ChIP-seq or ATAC-seq signals in the three clusters of ZMYM2 binding regions. (C) UCSC genome browser of the indicated ChIP-seq and ATAC-seq data on example genomic loci from clusters 1 and 2. See also Figure 2—figure supplement 1.
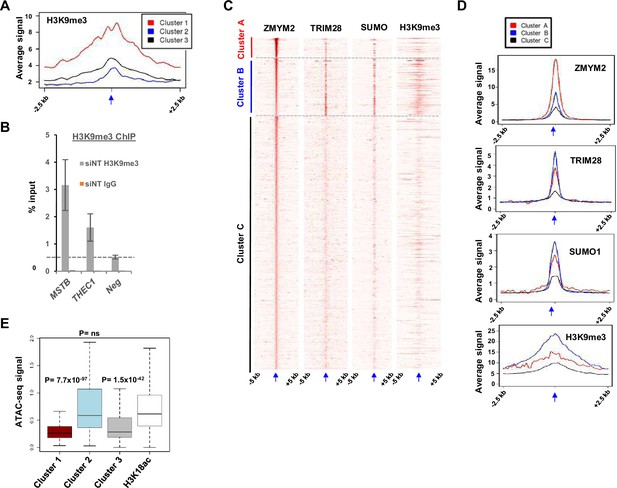
Molecular characterisation of ZMYM2 binding regions.
(A) Tag density plot of H3K9me3 ChIP-seq signal from H1 ESCs in the three clusters of ZMYM2 binding regions using ZMYM2 and TRIM28 binding regions for clustering. (B) ChIP-qPCR of H3K9me3 in U2OS cells at the indicated ZMYM2 bound loci or a negative control region (neg) not bound by ZMYM2. Signal from non-specific IgG is also shown at the same regions. Data are shown relative to input (n=3). (C) Heatmaps showing the signals of the indicated proteins or chromatin marks from ChIP-seq experiments in mouse ESCs cells plotted across a 10 kb region surrounding the centres (arrowed) of the ZMYM2 binding regions. Data were clustered to produce 3 clusters. (D) Tag density plots of the indicated ChIP-seq signals in the three clusters of ZMYM2 binding regions. (E) Boxplots showing the ATAC-seq signal at each of the ZMYM2 clusters and regions containing H3K18ac peaks. Significance values are shown relative to the signal at H3K18ac peaks.
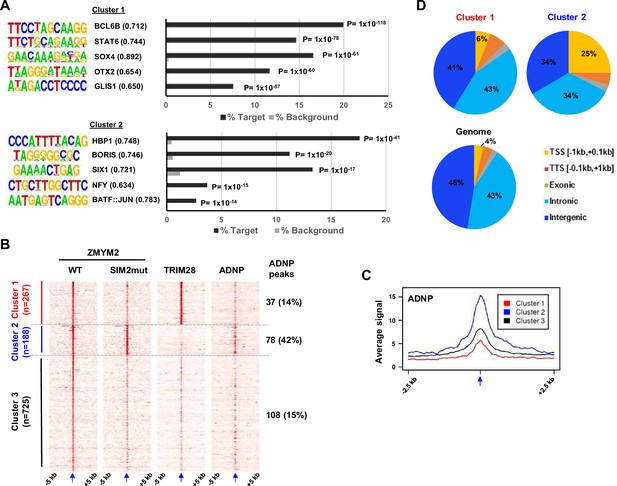
Characterisation of ZMYM2 chromatin binding regions.
(A) De novo motif analysis of cluster 1 and 2 regions. The top five most significantly enriched motifs are shown with motif similarity to the indicated protein shown in brackets. (B) Heatmaps showing the signals of the indicated proteins from ChIP-seq experiments in U2OS cells plotted across a 10 kb region surrounding the centres (arrowed) of the wild-type (WT) ZMYM2 binding regions. Clustering was retained from Figure 2A and ADNP signal superimposed on top of this. The number and percentage of ZMYM2 peaks in each cluster overlapping with ADNP2 peaks is shown on the right. (C) Tag density plot of ADNP ChIP-seq signal from U2OS cells across a 5 kb region surrounding the centres (arrowed) of the three clusters of ZMYM2 binding regions. (D) Distribution of binding regions among different genomic categories for clusters 1 and 2 and the entire genome. See also Figure 3—figure supplement 1.
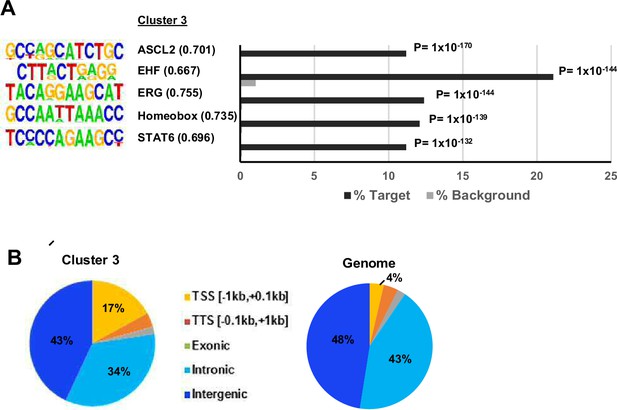
Genomic distributions and motif enrichments in ZMYM2 binding regions.
(A) De novo motif analysis of cluster 3 regions. The top five most significantly enriched motifs are shown with motif similarity to the indicated protein shown in brackets. (B) Distribution of binding regions among different genomic categories for cluster 3 and the entire genome.
-
Figure 3—figure supplement 1—source data 1
Raw unedited images of Co-immunoprecipitation analysis of TRIM28 with ZMYM2 (Figure 3—figure supplement 1A).
Resulting proteins were detected by immunoblotting (IB) with ZMYM2 antibody. This is a longer exposure of Figure 4A top panel to better visualize the input protein. The regions used for creating the final figure are boxed. Molecular weight marker sizes (kDa) are shown on the left.
- https://cdn.elifesciences.org/articles/86669/elife-86669-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Raw unedited images of Co-immunoprecipitation analysis of TRIM28 or SUMO2 with ZMYM2 (Figure 3—figure supplement 1B).
Resulting proteins were detected by immunoblotting (IB) with SUMO2 or TRIM28 antibodies. The regions used for creating the final figure are boxed. Molecular weight marker sizes (kDa) are shown on the left.
- https://cdn.elifesciences.org/articles/86669/elife-86669-fig3-figsupp1-data2-v1.zip
-
Figure 3—figure supplement 1—source data 3
Raw unedited images of co-immunoprecipitation (IP) analysis of endogenous TRIM28 with the indicated EGFP-tagged ZMYM2 proteins (Figure 3—figure supplement 1C, top).
IPs (top) were immunoblotted (IB) with the indicated antibodies. Resulting proteins were detected by immunoblotting (IB) with GFP or TRIM28 antibodies. The regions used for creating the final figure are boxed. Molecular weight marker sizes (kDa) are shown on the left.
- https://cdn.elifesciences.org/articles/86669/elife-86669-fig3-figsupp1-data3-v1.zip
-
Figure 3—figure supplement 1—source data 4
Raw unedited images of co-immunoprecipitation (IP) analysis of endogenous TRIM28 with the indicated EGFP-tagged ZMYM2 proteins (Figure 3—figure supplement 1C, bottom).
Input samples were immunoblotted (IB) with the indicated antibodies. The regions used for creating the final figure are boxed.
- https://cdn.elifesciences.org/articles/86669/elife-86669-fig3-figsupp1-data4-v1.zip
-
Figure 3—figure supplement 1—source data 5
Raw unedited images of co-immunoprecipitation analysis of TRIM28 with ZMYM3 (Figure 3—figure supplement 1F).
Immunoprecipitation (IP) was performed with ZMYM2 or control IgG antibody from U2OS cells in the presence of NEM and the resulting TRIM28 detected by immunoblotting (IB). 10% input is shown. The regions used for creating the final figure are boxed. Molecular weight marker sizes (kDa) are shown on the left.
- https://cdn.elifesciences.org/articles/86669/elife-86669-fig3-figsupp1-data5-v1.zip
-
Figure 3—figure supplement 1—source data 6
Raw unedited images of western blot analysis of lysates from U2OS cells treated with the indicated targeting or non-targeting (NT) siRNAs.
Lamin B (loading control), TRIM28 and ZMYM2 were detected by immunoblotting (IB) (Figure 3—figure supplement 1G). The regions used for creating the final figure are boxed. Molecular weight marker sizes (kDa) are shown on the left.
- https://cdn.elifesciences.org/articles/86669/elife-86669-fig3-figsupp1-data6-v1.zip
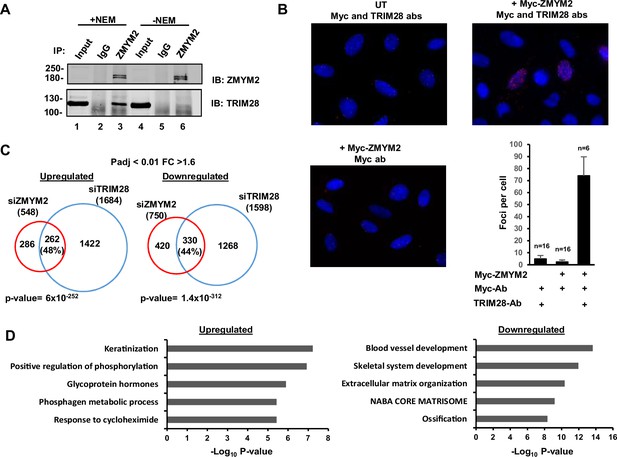
ZMYM2 interactions with TRIM28.
(A) Co-immunoprecipitation analysis of TRIM28 with ZMYM2. Immunoprecipitation (IP) was performed with ZMYM2 or control IgG antibody from U2OS cells and the resulting proteins detected by immunoblotting (IB) with the indicated antibodies. 10% input is shown (See Figure 4—figure supplement 1 for longer exposure). NEM was added to the extracts where indicated. (B) PLA assay of interactions between Myc-tagged ZMYM2 and endogenous TRIM28. Assays were carried out in U2OS cells transfected with a vector encoding Myc-ZMYM2 or left untransfected (UT). The addition of anti-Myc and -TRIM28 antibodies (ab) is indicated. Nuclei were stained with DAPI. Average numbers of foci per cell and numbers of cells are indicated (bottom). (C) Venn diagram showing the overlaps in upregulated (left) and downregulated (right) genes (Padj <0.05; fold change ≥1.6) from RNAseq analysis in U2OS cells treated with siRNAs against ZMYM2 or TRIM28. (D) Enriched GO terms of genes commonly upregulated and downregulated by ZMYM2 and TRIM28 depletion. See also Figure 4—figure supplement 1.
-
Figure 4—source data 1
Raw unedited images of Co-immunoprecipitation analysis of TRIM28 with ZMYM2 (Figure 4A).
Resulting proteins were detected by immunoblotting (IB) with ZMYM2 and TRIM28 antibodies. The regions used for creating the final figure are boxed. Molecular weight marker sizes (kDa) are shown on the left.
- https://cdn.elifesciences.org/articles/86669/elife-86669-fig4-data1-v1.zip
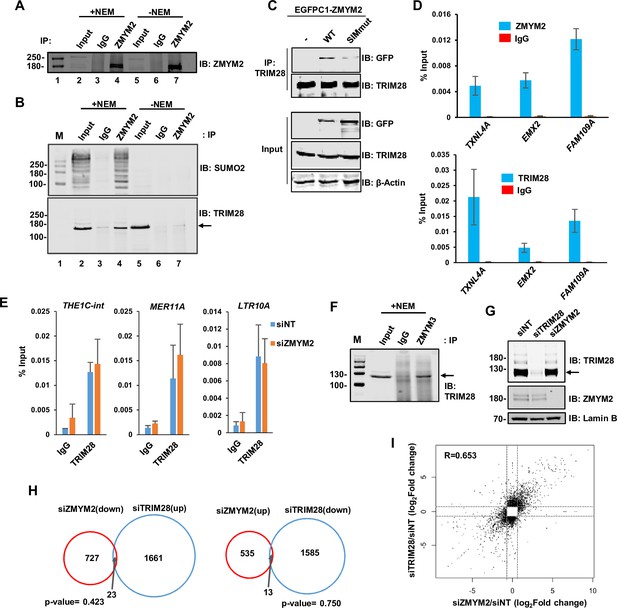
ZMYM2-TRIM28 interactions.
(A) Co-immunoprecipitation analysis of TRIM28 with ZMYM2. This is a longer exposure of Figure 4A top panel to better visualize the input protein. (B) Co-immunoprecipitation analysis of TRIM28 or SUMO2 with ZMYM2. Immunoprecipitation (IP) was performed with ZMYM2 or control IgG antibody from U2OS cells and the resulting proteins detected by immunoblotting (IB) with the indicated antibodies. 10% input is shown. NEM was added to the extracts where indicated. (C) Co-immunoprecipitation (IP) analysis of endogenous TRIM28 with the indicated EGFP-tagged ZMYM2 proteins. Inputs (bottom) and IPs (top) were immunoblotted (IB) with the indicated antibodies. (D) ChIP-qPCR of ZMYM2 (top) and TRIM28 (bottom) in U2OS cells at the indicated ZMYM2-bound loci. Signal from non-specific IgG is also shown at the same regions. Data are shown relative to input (n=3). (E) ChIP-qPCR of TRIM28 in U2OS cells following treatment with siZMYM2 or a non-targeting (NT) siRNA, at the indicated ZMYM2 bound loci. Signal from non-specific IgG is also shown at the same regions. Data are shown relative to input (n=2). (F) Co-immunoprecipitation analysis of TRIM28 with ZMYM3. Immunoprecipitation (IP) was performed with ZMYM3 or control IgG antibody from U2OS cells in the presence of NEM and the resulting TRIM28 detected by immunoblotting (IB). 10% input is shown. (G) Western blot analysis of lysates from U2OS cells treated with the indicated targeting or non-targeting (NT) siRNAs. Lamin B (loading control), TRIM28 and ZMYM2 were detected by immunoblotting (IB). (H) Venn diagrams showing overlaps in genes showing reciprocal directionality following ZMYM2 (left circle) or TRIM28 (right circle) depletion. Genes downregulated with siTRIM28 and upregulated with siZMYM2 (left) or upregulated with siTRIM28 and downregulated with siZMYM2 (right) are shown (fold change >1.6; Padj <0.01). (I) Scatterplot of significantly changing genes following ZMYM2 (x-axis) or TRIM28 (y-axis) depletion (fold change >1.6; Padj <0.01).
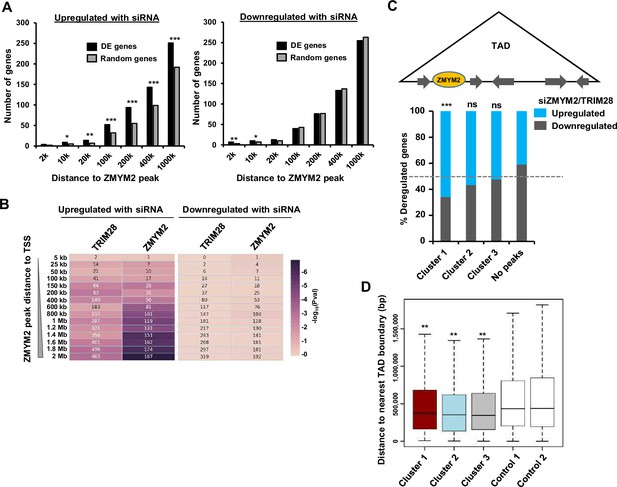
ZMYM2 location and gene expression.
(A) Numbers of differentially expressed genes following ZMYM2 depletion (left upregulated, right downregulated) whose TSS lies within the indicated distances of a ZMYM2 binding peak (black bars). Control sets of equal numbers of randomly selected genes are shown for comparison (grey bars showing the average of 10 datasets). p-values *=<0.05, **=<0.01, ***=<0.001. (B) Distance-dependent association of ZMYM2 binding regions with differentially regulated genes following depletion of ZMYM2 or TRIM28. The numbers in the boxes are the number of genes among the input gene sets (x-axis) that overlap with the ZMYM2 peaks (cluster 1 peaks only) at the indicated distances to TSS (y-axis) and the colour shows − log10 of p-value (Hypergeometric test). (C) Relative proportion of genes commonly up- or down-regulated following ZMYM2 or TRIM28 depletion in TADs which also contain ZMYM2 peaks from the indicated clusters or have no ZMYM2 peak in the same TADs. Significance relative to regions containing no peaks is shown p-value, ***=<0.001; ns = non-significant. (D) Boxplots of the relative distance of ZMYM2 binding regions from each of the clusters from TAD boundaries compared to two different control sets of randomly selected regions (n=2360). Statistical significance of cluster 1–3 distances compared to each of the control regions is shown (**=p-value<0.05 in all cases; student t-test). See also Figure 5—figure supplement 1.

ZMYM2 location and gene expression.
(A) Overlap between the closest genes to ZMYM2 binding regions from cluster 1 and the intersection of differentially expressed genes following ZMYM2 or TRIM28 depletion. (B) Relative proportion of genes up- or down-regulated following either ZMYM2 (top) or TRIM28 (bottom) depletion in TADs which also contain ZMYM2 peaks from the indicated clusters or have no ZMYM2 peaks in the same TADs. (C) Frequency distribution of the number of TADs containing a gene upregulated (fold change >1.6; Padj <0.01) following ZMYM2 depletion. A total of 10,000 iterations were performed by randomly selecting 216 TADs across all 3062 TADs. The observed number of TADs containing an upregulated gene (42) from the 216 TADs containing a cluster 1 ZMYM2 peak is shown (p-value = 0.0002).
-
Figure 5—figure supplement 1—source data 1
Raw unedited images of western blot illustrating the knockdown efficiencies of ZMYM2 in the RT-qPCR experiments.
β-actin is shown as a loading control (Figure 5—figure supplement 1A). The regions used for creating the final figure are boxed. Molecular weight marker sizes (kDa) are shown on the left.
- https://cdn.elifesciences.org/articles/86669/elife-86669-fig5-figsupp1-data1-v1.zip
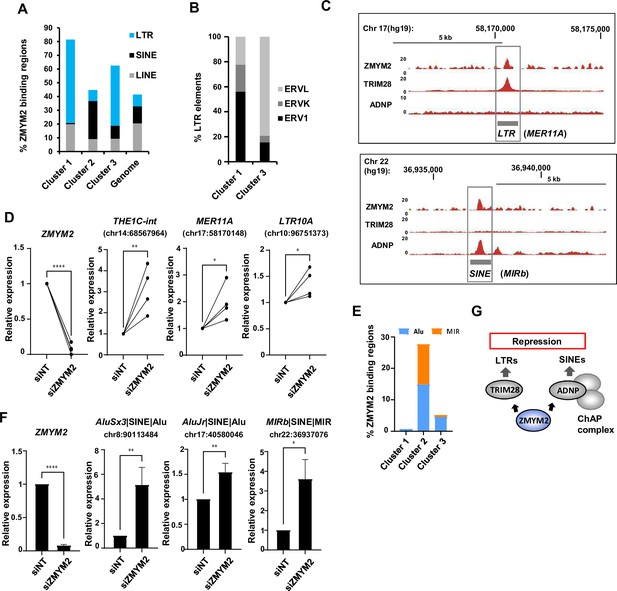
ZMYM2 functionally associates with ERV repetitive elements.
(A) Percentage of the ZMYM2 binding sites in each of the indicated clusters containing each of the indicated classes of retrotransposon elements. The genome-wide proportion of genomic regions containing each type of the retrotransposon elements is also shown. (B) Proportions of LTR subclasses in cluster 1 and cluster 3 ZMYM2 binding regions. (C) UCSC genome browser view of a MER11A ERV1 LTR element located upstream of the HEATR6 locus, illustrating co-binding of ZMYM2 and TRIM28 (top) and a MIR SINE element located upstream of the EIF3D locus, illustrating co-binding of ZMYM2 and ADNP. (D) RT-qPCR analysis of expression of ZMYM2 and the indicated LTR elements following ZMYM2 depletion or control non-targeting (NT) siRNA treatment. Individual paired experiments are shown (n=4; p-values *=<0.05,**=<0.01, ****=<0.0001). (E) Proportions of SINE subclasses in cluster 1–3 ZMYM2 binding regions. (F) RT-qPCR analysis of expression of ZMYM2 and the indicated SINE elements following ZMYM2 depletion or control non-targeting (NT) siRNA treatment (n=3; unpaired T-test p-values *=<0.05, **=<0.01, ****=<0.0001). (G) Model illustrating the two distinct complexes through which ZMYM2 functions on chromatin to control retrotransposon expression. See also Figure 6—figure supplement 1.
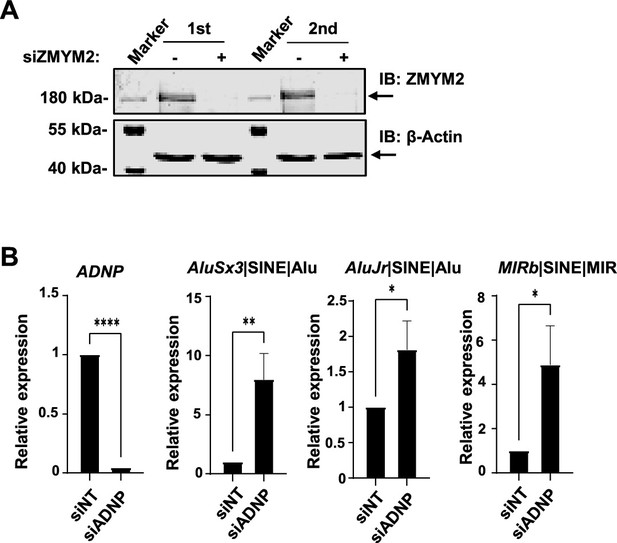
ZMYM2 containing complexes regulate retrotransposons.
(A) Western blot illustrating the knockdown efficiencies of ZMYM2 in the RT-qPCR experiments. b-actin is shown as a loading control and sizes of molecular weight markers are indicated. (B) RT-qPCR analysis of expression of ADNP and the indicated SINE elements following ADNP depletion or control non-targeting (NT) siRNA treatment. Individual paired experiments are shown (n=3; p-values *=<0.05, **=<0.01, ****=<0.0001).
Additional files
-
Supplementary file 1
List of antibodies.
- https://cdn.elifesciences.org/articles/86669/elife-86669-supp1-v1.docx
-
Supplementary file 2
Sequencing statistics.
- https://cdn.elifesciences.org/articles/86669/elife-86669-supp2-v1.xlsx
-
Supplementary file 3
RIME analysis of the ZMYM2 interactome.
- https://cdn.elifesciences.org/articles/86669/elife-86669-supp3-v1.xlsx
-
Supplementary file 4
RNAseq analysis of gene expression changes following ADNP, ZMYM2 and TRIM28 depletion.
- https://cdn.elifesciences.org/articles/86669/elife-86669-supp4-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/86669/elife-86669-mdarchecklist1-v1.docx
-
Source data 1
Original western blots used to create cropped figures shown in thje manuscipt.
- https://cdn.elifesciences.org/articles/86669/elife-86669-data1-v1.pptx





