A MSTNDel73C mutation with FGF5 knockout sheep by CRISPR/Cas9 promotes skeletal muscle myofiber hyperplasia
Figures
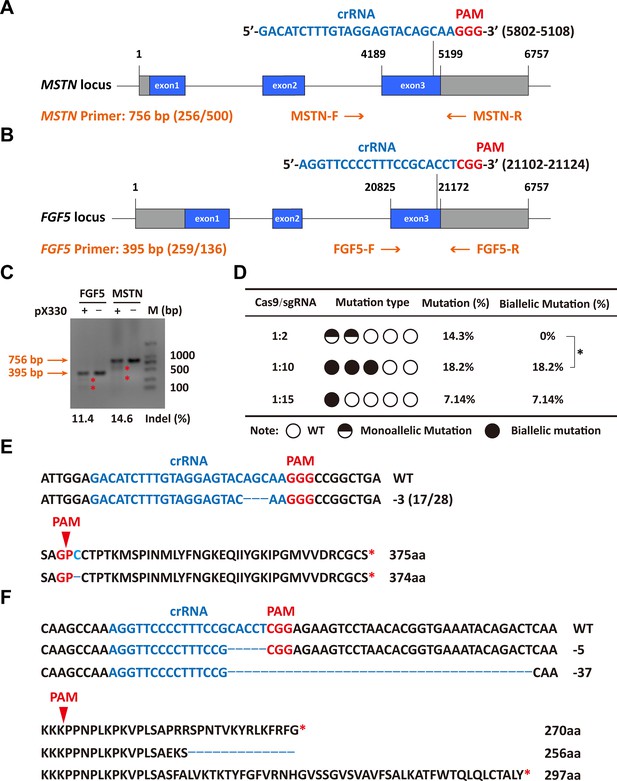
Efficient generation of sheep carrying biallelic mutations in dual gene via the CRISPR/Cas9 system.
(A) Schematic of sgRNAs specific to exon 3 of the sheep MSTN locus. The crRNA sequences are highlighted in blue typeface and the PAM in red. (B) Schematic of sgRNAs specific to exon 3 of the sheep FGF5 locus. The crRNA sequences are highlighted in blue typeface and the PAM in red. (C) T7EI assay for sgRNAs of MSTN and FGF5 in sheep fetal fibroblasts. The cleavage bands are marked with an red asterisk (*) and the indel frequencies were calculated using the expected fragments. (D) Summary of the generation of sheep carrying biallelic mutations in dual gene via zygote injection of Cas9 mRNA/sgRNAs. Biallelic mutation rate was statistically analyzed using chi square test. *p<0.05. (E) Analysis of genome sequence and amino acid sequence of MSTN-modified sheep. The location of sgRNA and PAM are highlighted in blue and red, respectively. The deletions are indicated by a dashed line (-). (F) Analysis of genome sequence and amino acid sequence of FGF5-modified sheep. The location of sgRNA and PAM are highlighted in blue and red, respectively. The deletions are indicated by a dashed line (-).
-
Figure 1—source data 1
Uncropped and labeled gels for Figure 1C.
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig1-data1-v1.zip
-
Figure 1—source data 2
Raw unedited gels for Figure 1C.
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig1-data2-v1.zip
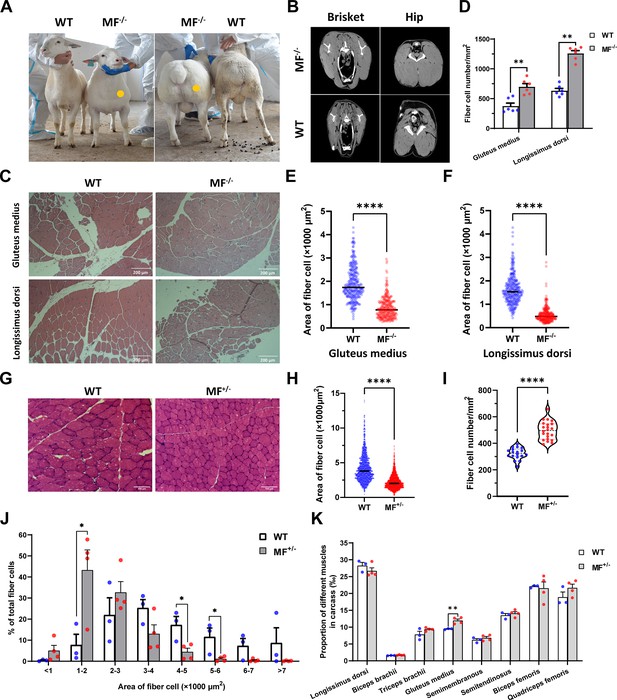
The MSTNDel73C mutation with FGF5 knockout sheep highlights a dominant ‘double-muscle’ phenotype and muscle fiber hyperplasia.
(A) The 6-month-old WT and MF-/- sheep. The genome-edited sheep displayed an obvious ‘double-muscle’ phenotype compared with the WT. (B) The CT scanning image of the brisket and hip of WT and MF-/- sheep. (C) HE sections of gluteus medius and longissimus dorsi of WT and MF-/- sheep. Scale bar 200 μm. (D) Quantification of muscle fibre cell number of per unit area in WT (n=3) and MF-/- (n=1) sheep. All data points were shown. (E–F) Quantification of muscle fibre cell area of gluteus medius and longissimus dorsi in WT (n=3) and MF-/- (n=1) sheep. All data points were shown. (G) HE sections of gluteus medius in WT and MF+/- sheep. Scale bar 100 μm. (H) Quantification of muscle fibre cell area of gluteus medius in WT (n=3) and MF+/- (n=4) sheep. (I) Quantification of muscle fibre cell number of per unit area in WT (n=3) and MF+/- (n=4) sheep. (J) The percentage of cross-sectional area of different size muscle fibers. (K) The proportion of different muscles in carcass in WT (n=3) and MF+/- (n=4) sheep. Data: mean ± SEM. Unpaired student’s t-test were used for statistical analysis after the equal variance test, otherwise the t-test with Welch’s correction were performed. *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001.
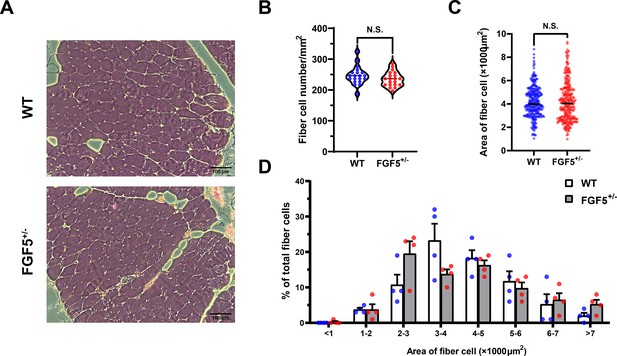
FGF5 mutation does not affect muscle fiber size.
(A) HE sections of gluteus medius in WT and FGF5+/- sheep. Scale bar +/- μm. (B) Quantification of muscle fibre cell area of gluteus medius in WT and FGF5+/- sheep. (C) Quantification of muscle fibre cell number of per unit area in WT and FGF5+/- sheep. (D) The percentage of cross-sectional area of different size muscle fibers. Data: mean ± SEM. Unpaired student’s t-test was used for statistical analysis. All student’s t-test were performed after the equal variance test, otherwise the t-test with Welch’s correction were used.
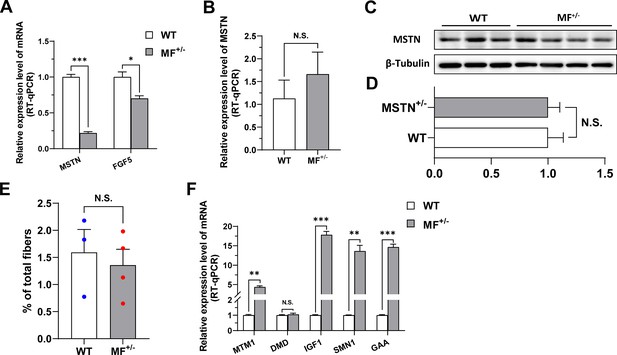
The MSTNDel73C mutation with FGF5 knockout has no potential effect on MSTN expression and muscular dystrophy.
(A) The mRNA expression levels of MSTN and FGF5 of gluteus medius in WT (n=4) and MF+/- (n=4) sheep at 3-month-old. (B) MSTN mRNA expression level of gluteus medius in WT (n=3) and MF+/- (n=4) sheep. (C–D) MSTN protein expression level of gluteus medius in WT (n=3) and MF+/- (n=4) sheep. (E) The proportion of centrally nucleated myofibres between WT (n=3) and MF+/- (n=4) sheep. (F) The mRNA expression of muscular dystrophy related genes between WT and MF+/- sheep (n=4). Data: mean ± SEM. Unpaired student’s t-test was used for statistical analysis. All student’s t-test were performed after the equal variance test, otherwise the t-test with Welch’s correction were used. *p<0.05, **p<0.01, and ***p<0.001.
-
Figure 2—figure supplement 2—source data 1
Uncropped and labeled blots for Figure 2—figure supplement 2C.
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig2-figsupp2-data1-v1.zip
-
Figure 2—figure supplement 2—source data 2
Raw unedited blots for Figure 2—figure supplement 2C.
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig2-figsupp2-data2-v1.zip
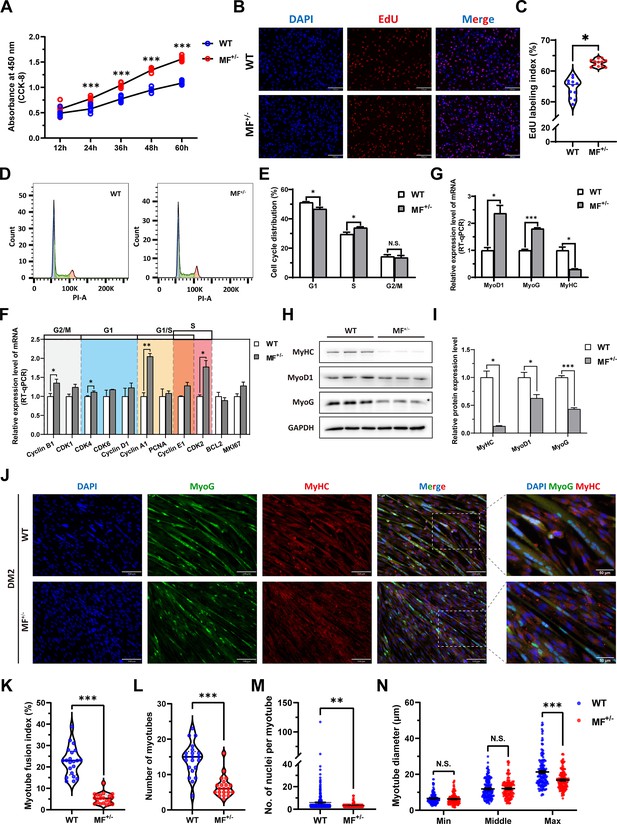
The MSTNDel73C mutation with FGF5 knockout promote proliferation and inhibit differentiation of skeletal muscle satellite cells.
(A) The number of cells was detected by CCK-8 at 12 hr, 24 hr, 36 hr, 48 hr, and 60 hr in GM (n=7–8 per group). (B–C) EdU assay showed that the number of EdU positive cells and EdU labeling index were significantly increased in MF+/- cells (n=3). Scale bar 130 μm. All data points were shown. (D–E) PI staining to detect cell cycle and showed a significant reduce in the proportion of G1 phase and a significant increase in the proportion of S phase in MF+/- cells (n=4). (F) The mRNA expression levels of cell cycle marker genes and cell proliferation marker genes (n=3). (G) The mRNA expression levels of myogenic differentiation marker genes MyoG, MyoD1, and MyHC (n=3). (H–I) The protein expression levels of myogenic differentiation marker genes MyoG, MyoD1, and MyHC (n=3). (J) The MyoG and MyHC immunofluorescence staining of myotubes in DM2. Scale bar 130 μm. (K) The myotube fusion index, which was represented by the number of cell nuclei in myotubes/total cell nuclei (n=3). All data points were shown. (L) The number of myotubes, which was the number of all myotubes in the field of view (n=3). All data points were shown. (M) The number of nuclei per myotube (n=3). All data points were shown. (N) The myotube diameter (n=3). To reflect the myotube diameter as accurately as possible, the vertical line at the thinnest position of the myotube is taken as the minimum measured (Min), the mid-perpendicular line of the long myotube axis is taken as the middle measured (Middle), and the vertical line at the widest position of the myotube is taken as the maximum measured (Max). All data points were shown. Data: mean ± SEM. Unpaired student’s t-test and chi square test were used for statistical analysis. All student’s t-test were performed after the equal variance test, otherwise the t-test with Welch’s correction were used. *p<0.05, **p<0.01, and ***p<0.001.
-
Figure 3—source data 1
Uncropped and labeled blots for Figure 3H.
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig3-data1-v1.zip
-
Figure 3—source data 2
Raw unedited blots for Figure 3H.
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig3-data2-v1.zip
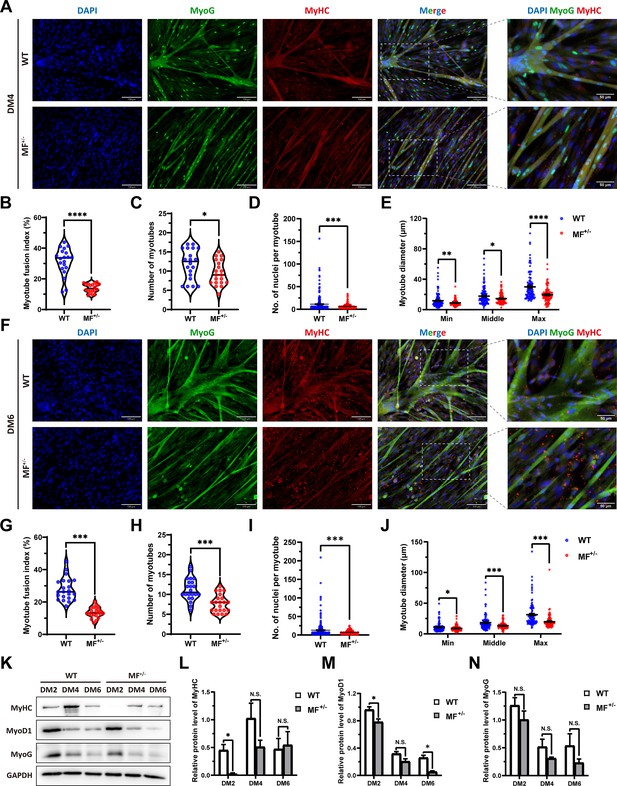
The myogenic differentiation ability of MF+/- cells was continuously inhibited.
(A) The MyoG and MyHC immunofluorescence staining of myotubes at DM4. Scale bar 130 μm. (B–E) The myotube fusion index, number of myotubes, number of nuclei per myotube and the myotube diameter at DM4 (n=3). (F) The MyoG and MyHC immunofluorescence staining of myotubes at DM6. Scale bar 130 μm. (G–J) The myotube fusion index, number of myotubes, number of nuclei per myotube and the myotube diameter at DM6 (n=3). (K–N) The protein expression levels of myogenic differentiation markers during myogenic differentiation between WT and MF+/- cells (n=2–3). Data: mean ± SEM. Unpaired student’s t-test and chi square test were used for statistical analysis. All student’s t-test were performed after the equal variance test, otherwise the t-test with Welch’s correction were used. *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001.
-
Figure 3—figure supplement 1—source data 1
Uncropped and labeled blots for Figure 3—figure supplement 1K.
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Raw unedited blots for Figure 3—figure supplement 1K.
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig3-figsupp1-data2-v1.zip
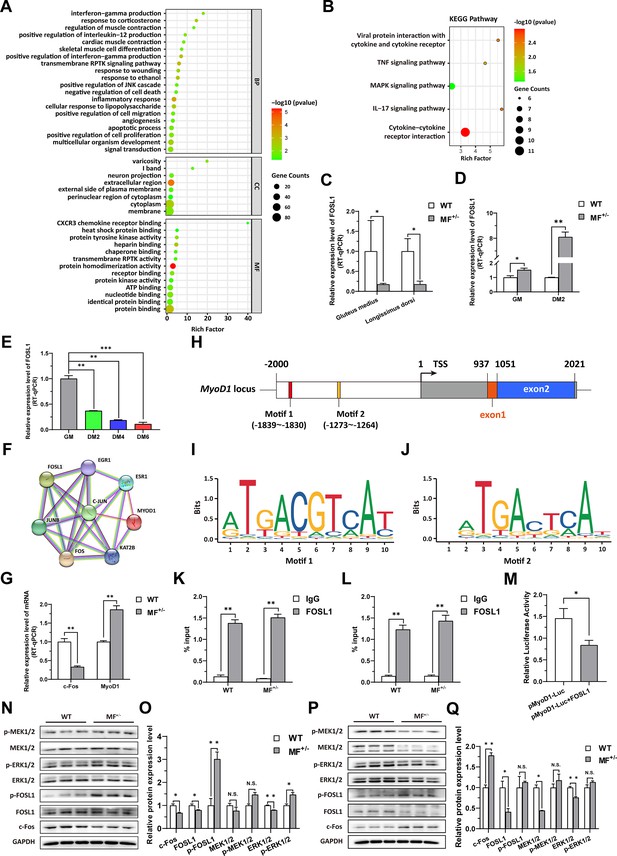
The MSTNDel73C mutation with FGF5 knockout contributes to muscle phenotype via MEK-ERK-FOSL1 axis.
(A) Go enrichment analysis of DEGs. Among them, the top 20 entries with significant enrichment are listed in biological process (BP). CC, cellular component; MF, molecular function. (B) KEGG enrichment analysis of DEGs. (C) The mRNA expression level of FOSL1 both at gluteus medius and longissimus dorsi in WT (n=3) and MF+/- (n=4) sheep. (D) The mRNA expression level of FOSL1 both at GM and DM2 in WT and MF+/- cells (n=3). (E) The expression level of FOSL1 mRNA during myogenic differentiation (n=3). (F) The protein-protein interaction (PPI) analysis of FOSL1, c-Fos and MyoD1. (G) The mRNA expression level of c-Fos and MyoD1 at GM in WT and MF+/- myoblasts (n=3). (H) Schematic diagram of MyoD1 gene body, promoter region and binding sites. (I–J) FOSL1 recognition motif in the MyoD1 promoter region. (K) FOSL1 ChIP-qPCR of motif 1 recognition region (n=3). (L) FOSL1 ChIP-qPCR of motif 2 recognition region (n=3). (M) Dual luciferase assay for the effect of FOSL1 on MyoD1 promoter activity (n=4). (N) Western blot of FOSL1, c-Fos, and key kinases of MAPK signaling pathways at GM. (O) Quantification of protein expression of FOSL1, c-Fos, and key kinases of MAPK signaling pathways at GM (n=3). (P) Western blot of FOSL1, c-Fos, and key kinases of MAPK signaling pathways at DM2. (Q) Quantification of protein expression of FOSL1, c-Fos, and key kinases of MAPK signaling pathways at DM2 (n=3). Data: mean ± SEM. Unpaired student’s t-test was used for statistical analysis. All student’s t-test were performed after the equal variance test, otherwise the t-test with Welch’s correction were used. *p<0.05, **p<0.01, and ***p<0.001.
-
Figure 4—source data 1
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig4-data1-v1.zip
-
Figure 4—source data 2
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig4-data2-v1.zip
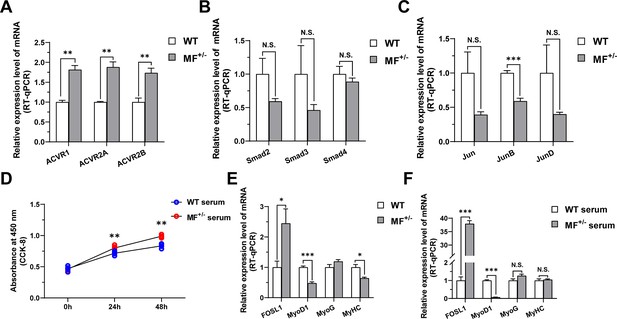
The effects of MSTN signaling pathway and MF+/- serum on proliferation and differentiation of skeletal muscle satellite cells.
(A) The mRNA expression levels of type I and type II receptors of MSTN between WT (n=3) and MF+/- (n=4) sheep gluteus medius. (B) The mRNA expression levels of the Smad family downstream of MSTN between WT (n=3) and MF+/- (n=4) sheep gluteus medius. (C) The mRNA expression levels of the Jun family downstream of MSTN between WT (n=3) and MF+/- (n=4) sheep gluteus medius. (D) The number of cells detected by CCK-8 at 0 hr, 24 hr, and 48 hr after culturing cells with serum from WT sheep and MF+/- sheep (n=4–5). (E) The mRNA expression levels of FOSL1 and myogenic differentiation markers between WT and MF+/- sheep skeletal muscle satellite cells cultured and induced differentiation in sheep serum (n=4). (F) The mRNA expression levels of FOSL1 and myogenic differentiation markers in skeletal muscle satellite cells cultured and induced differentiation by serum from WT and MF+/- sheep (n=4). Data: mean ± SEM. Unpaired student’s t-test and chi square test were used for statistical analysis. All student’s t-test were performed after the equal variance test, otherwise the t-test with Welch’s correction were used. *p<0.05, **p<0.01, and ***p<0.001.
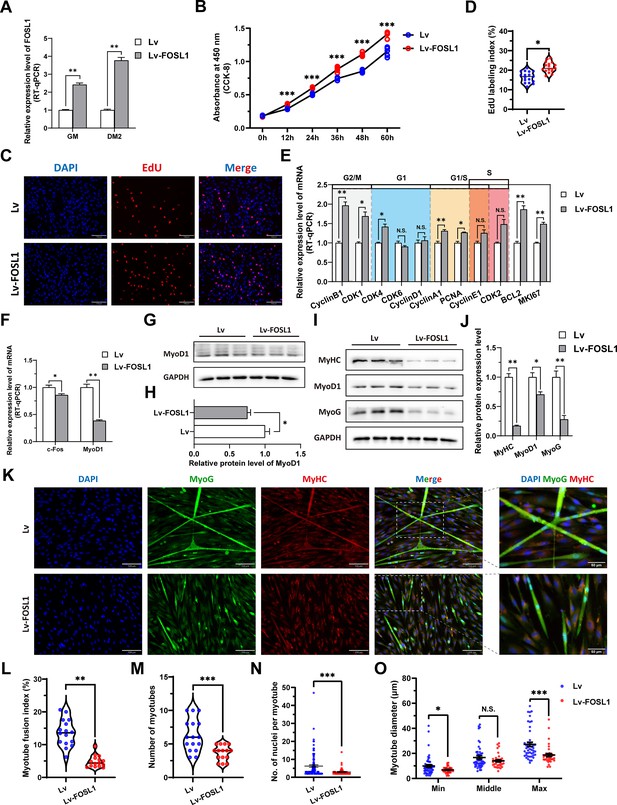
The overexpression of FOSL1 promotes proliferation and inhibits differentiation of skeletal muscle satellite cells.
(A) The mRNA expression level of FOSL1 at GM and DM2 after lentivirus infection (n=3). (B) The number of cells detected by CCK-8 at 0 hr, 12 hr, 24 hr, 36 hr, 48 hr, and 60 hr after infection with lentivirus (n=4–6). (C–D) EdU assay showed that the number of EdU-positive cells and EdU labeling index were significantly increased after infection with lentivirus (n=3). Scale bar 130 μm. All data points were shown. (E) The mRNA expression levels of cell cycle marker genes and cell proliferation marker genes (n=3). (F) The mRNA expression levels of c-Fos and MyoD1 at GM after overexpression of FOSL1 (n=3). (G–H) The protein expression levels of MyoD1 at GM after overexpression of FOSL1 (n=3). (I–J) The protein expression levels of myogenic differentiation marker genes MyoD1, MyoG and MyHC at DM2 after overexpression of FOSL1 (n=3). (K) The MyoG and MyHC immunofluorescence staining of myotubes at DM2 after overexpression of FOSL1. Scale bar 130 μm. (L–O) The myotube fusion index, number of myotubes, number of nuclei per myotube and the myotube diameter at DM2 after overexpression of FOSL1 (n=3). All data points were shown. Data: mean ± SEM. Unpaired student’s t-test and chi square test were used for statistical analysis. All student’s t-test were performed after the equal variance test, otherwise the t-test with Welch’s correction were used. *p<0.05, **p<0.01, and ***p<0.001.
-
Figure 5—source data 1
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig5-data1-v1.zip
-
Figure 5—source data 2
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig5-data2-v1.zip
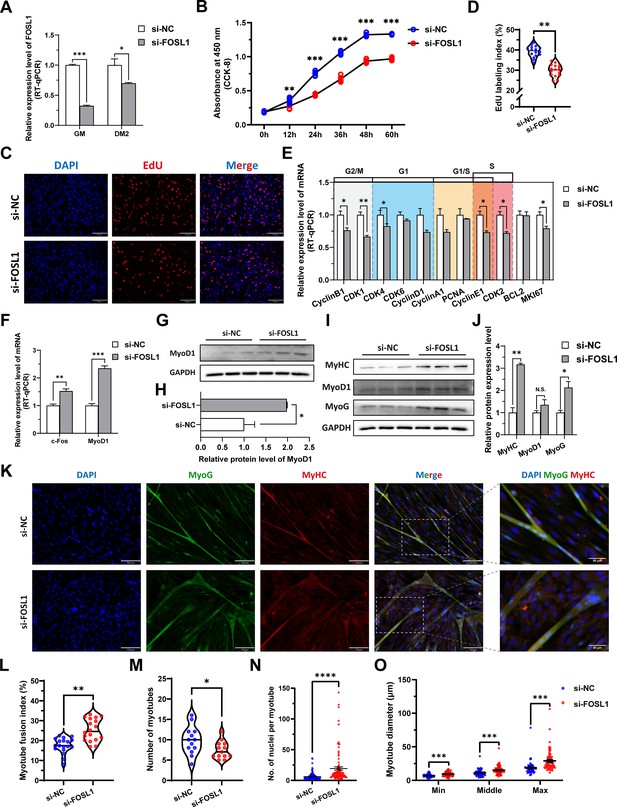
The inhibition of FOSL1 suppresses proliferation and promotes differentiation of skeletal muscle satellite cells.
(A) The mRNA expression level of FOSL1 at GM and DM2 after inhibiting FOSL1 (n=3). (B) The number of cells detected by CCK-8 at 0 hr, 12 hr, 24 hr, 36 hr, 48 hr, and 60 hr after inhibiting FOSL1 (n=4–6). (C–D) EdU assay showed that the number of EdU-positive cells and EdU labeling index were significantly decreased after inhibiting FOSL1 (n=3). Scale bar 130 μm. All data points were shown. (E) The mRNA expression levels of cell cycle marker genes and cell proliferation marker genes (n=3). (F) The mRNA expression levels of c-Fos and MyoD1 at GM after inhibiting FOSL1 (n=3). (G–H) The protein expression levels of MyoD1 at GM after inhibiting FOSL1 (n=3). (I–J) The protein expression levels of myogenic differentiation marker genes MyoD1, MyoG and MyHC at DM2 after inhibiting FOSL1 (n=3). (K) The MyoG and MyHC immunofluorescence staining of myotubes at DM2 after inhibiting FOSL1. Scale bar 130 μm. (L–O) The myotube fusion index, number of myotubes, number of nuclei per myotube and the myotube diameter at DM2 after overexpression of FOSL1 (n=3). All data points were shown. Data: mean ± SEM. Unpaired student’s t-test and chi square test were used for statistical analysis. All student’s t-test were performed after the equal variance test, otherwise the t-test with Welch’s correction were used. *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001.
-
Figure 6—source data 1
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig6-data1-v1.zip
-
Figure 6—source data 2
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig6-data2-v1.zip
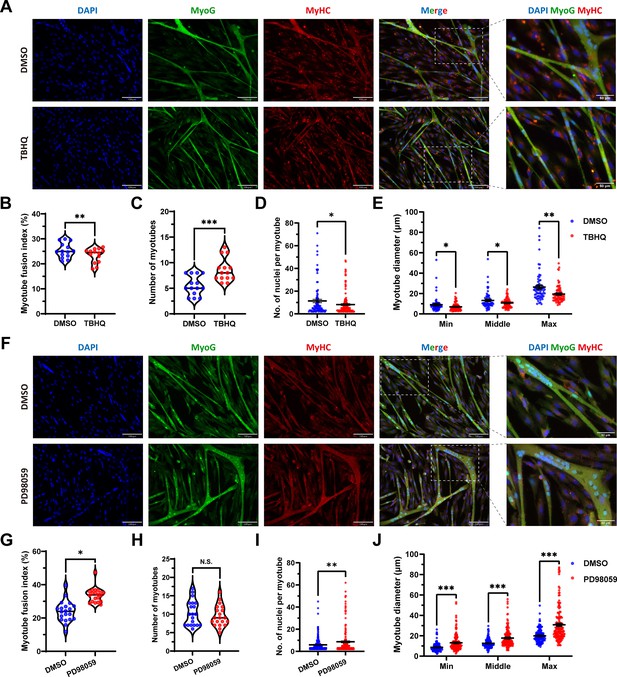
FOSL1 activity is a key regulator of myogenic differentiation and muscle myofiber hyperplasia.
(A) Immunofluorescence staining of myogenic differentiation markers MyoG and MyHC in sheep skeletal muscle satellite cells at DM2 after addition of 20 μM TBHQ. Scale bar 130 μm or 50 μm. (B–E) The myotube fusion index, number of myotubes, number of nuclei per myotube and the myotube diameter at DM2 after addition of 20 μM TBHQ (n=3). All data points were shown. (F) Immunofluorescence staining of myogenic differentiation markers MyoG and MyHC in sheep skeletal muscle satellite cells at DM2 after addition of 1 μM PD98059. Scale bar 130 μm or 50 μm. (G–J) The myotube fusion index, number of myotubes, number of nuclei per myotube and the myotube diameter at DM2 after addition of 1 μM PD98059 (n=3). All data points were shown. Data: mean ± SEM. Unpaired student’s t-test and chi square test were used for statistical analysis. All student’s t-test were performed after the equal variance test, otherwise the t-test with Welch’s correction were used. *p<0.05, **p<0.01, and ***p<0.001.
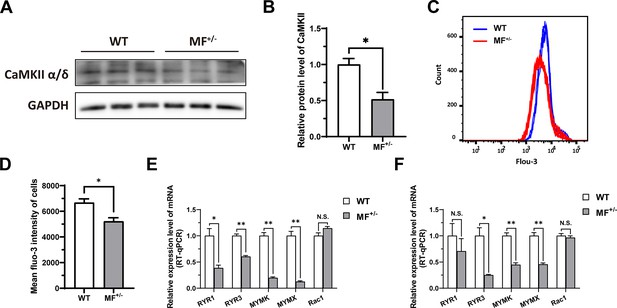
The MSTNDel73C mutation with FGF5 knockout inhibit calcium-dependent transcription signal pathway.
(A–B) The protein expression level of CaMKII α/δ between WT and MF+/- cells at GM (n=3). (C) Distribution of intracellular Ca2+ signals between WT and MF+/- cells at GM. (D) Average intracellular Ca2+ fluorescence intensity between WT and MF+/- cells at GM (n=4). (E) The mRNA expression levels of Ca2+ channels and myoblast fusion-related genes at GM (n=3). (F) The mRNA expression levels of Ca2+ channels and myoblast fusion-related genes at DM2 (n=3). Data: mean ± SEM. Unpaired student’s t-test was used for statistical analysis. All student’s t-test were performed after the equal variance test, otherwise the t-test with Welch’s correction were used. *p<0.05, **p<0.01.
-
Figure 8—source data 1
Uncropped and labeled blots for Figure 8A.
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig8-data1-v1.zip
-
Figure 8—source data 2
Raw unedited blots for Figure 8A.
- https://cdn.elifesciences.org/articles/86827/elife-86827-fig8-data2-v1.zip
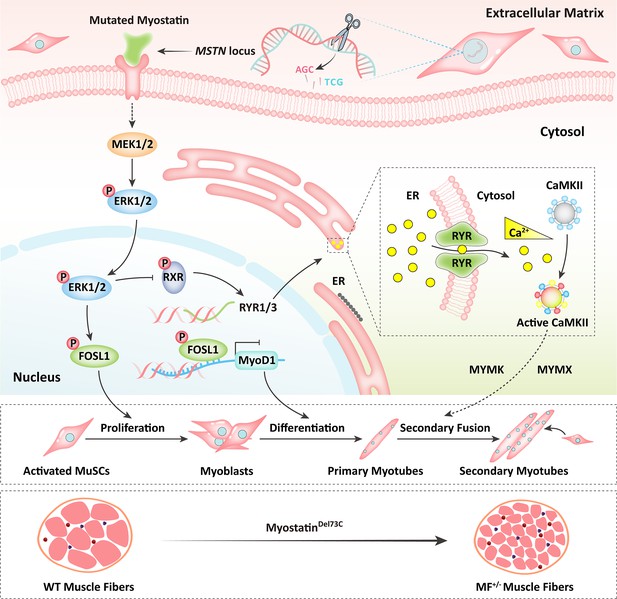
Schematic illustration of the regulation of muscle phenotypes by MSTNDel73C mutation with FGF5 knockout.
The MSTNDel73C mutation with FGF5 knockout mediated the activation of FOSL1 via MEK-ERK-FOSL1 axis. The activated FOSL1 promotes skeletal muscle satellite cell proliferation and inhibits myogenic differentiation by inhibiting the expression of MyoD1, and resulting in fusion to form smaller myotubes. In addition, activated ERK1/2 may inhibit the secondary fusion of myotubes by Ca2+-dependent CaMKII activation pathway, leading to myoblasts fusion to form smaller myotubes.
Additional files
-
Supplementary file 1
Production and slaughter information of MSTNDel73C mutation with FGF5 knockout sheep.
(A) Summary of generation of sheep carrying biallelic mutations in dual genes via the CRISPR/Cas9 system. (B) Muscle weight of different parts in WT and MF+/- sheep (g). (C) The slaughter traits of muscles in WT and MF+/- sheep. (D) Meat quality of longissimus dorsi in WT and MF+/- sheep. (E) Shearing force of different parts in WT and MF+/- sheep (N). (F) Amino acid content of longissimus dorsi in WT and MF+/- sheep (%, DM basis).
- https://cdn.elifesciences.org/articles/86827/elife-86827-supp1-v1.docx
-
Supplementary file 2
The information of primers sequence, siRNA sequence and antibody.
(A) Primers sequences of gene cloning. (B) The sequences of siRNA. (C) All primers of PCR and RT-qPCR. (D) The antibodies information.
- https://cdn.elifesciences.org/articles/86827/elife-86827-supp2-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/86827/elife-86827-mdarchecklist1-v1.docx





