Leveraging genetic diversity to identify small molecules that reverse mouse skeletal muscle insulin resistance
Figures
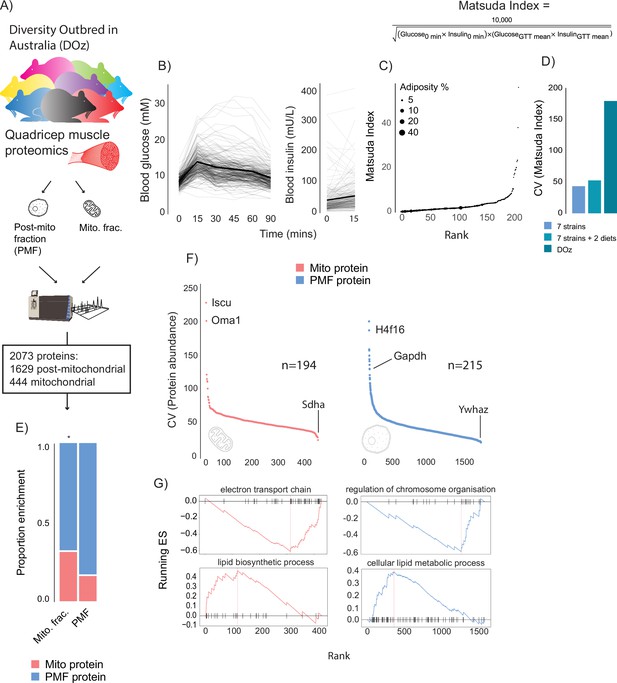
Metabolic and proteomic diversity of Diversity Outbred in Oz (DOz) mice.
(A) Schematic of metabolic phenotyping and quadricep proteomics in chow-fed DOz mice. (B) Blood glucose and insulin levels during a glucose tolerance test (GTT). (C) Whole-body insulin sensitivity (Matsuda Index, formula shown above) and adiposity of DOz mice (n = 215). (D) Comparison of coefficient of variation (CV) of insulin of Matsuda Index across inbred strains and diets versus chow-fed DOz mice. (E) Relative enrichment of mitochondrial (Mito) proteins in mitochondrial fraction and post-mitochondrial fraction (PMF) of quadricep proteomes. (F) Relative protein CV across mitochondrial and post-mitochondrial quadricep fractions. (G) Biological pathways enriched in mitochondrial and post-mitochondrial quadricep fractions, running enrichment score for a given pathway (ES) on y-axis and proteins ranked by CV on x-axis. Significance testing was performed by chi-square test. * indicates a significant difference p<0.01.
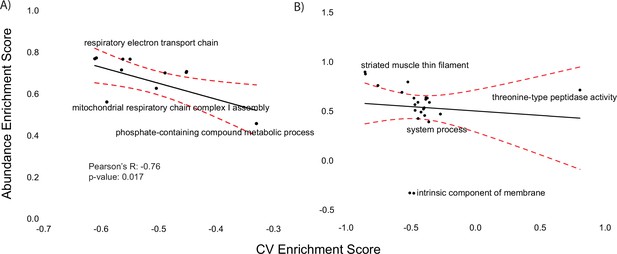
Assessment of variation-based enrichment analysis Diversity Outbred in Oz (DOz) muscle proteomics.
(A–, B) Abundance (y-axis) and coefficient of variation-based (x-axis) Gene Ontology enrichment scores for mitochondrial (A) and post-mitochondrial fraction (PMF) (B) proteomes. Dashed red line indicates 95% confidence interval for linear regression analysis.
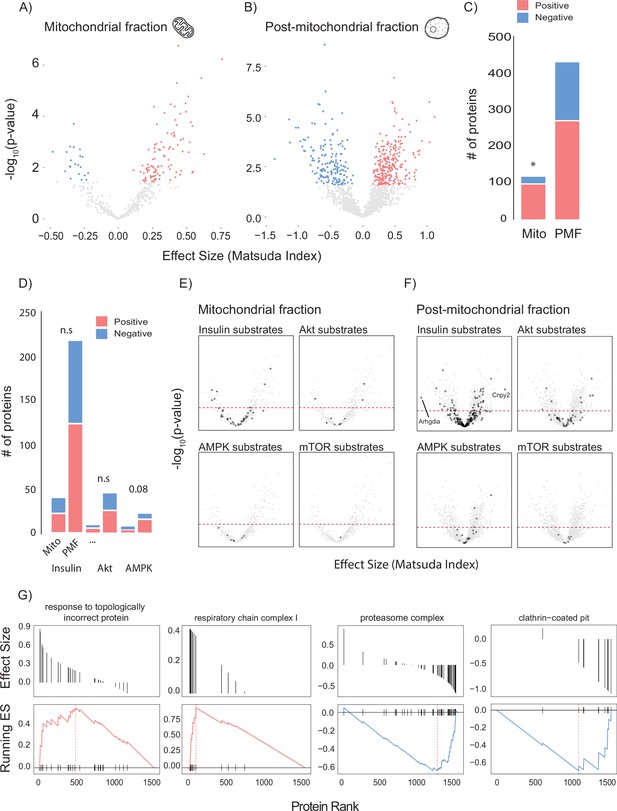
Linear modelling of quadricep proteome and whole-body insulin sensitivity.
(A, B) Volcano plot with Matsuda Index effect sizes (x-axis) and significance (y-axis) for mitochondrial (A) and post-mitochondrial (B) quadricep proteins using a linear model with adiposity as a covariate. Significant proteins with positive and negative effect sizes are indicated in red and blue, respectively. (C) Comparison of positively and negatively associated proteins between fractions. (D) Number of proteins identified in each fraction with known roles in insulin or AMPK signalling. (E, F) Volcano plot shown in (A) with mitochondrial (E) and post-mitochondrial fraction (F) proteins shown in black that have documented roles in indicated signalling pathways. Adjusted p-value threshold is indicated (red dotted line). (G) Pathways enriched for proteins which positively and negative associate with Matsuda Index. Effect sizes of proteins within a given pathway and running enrichment score (ES) for a that pathway on y-axis, and proteins ranked by CV on x-axis. Proteins ranked by Matsuda Index effect size on x-axis. Linear modelling was performed using a Gaussian distribution with q-value adjustment of p-values. Enrichment tests between fractions were performed by chi-square test. * indicates a significant difference p<0.01.
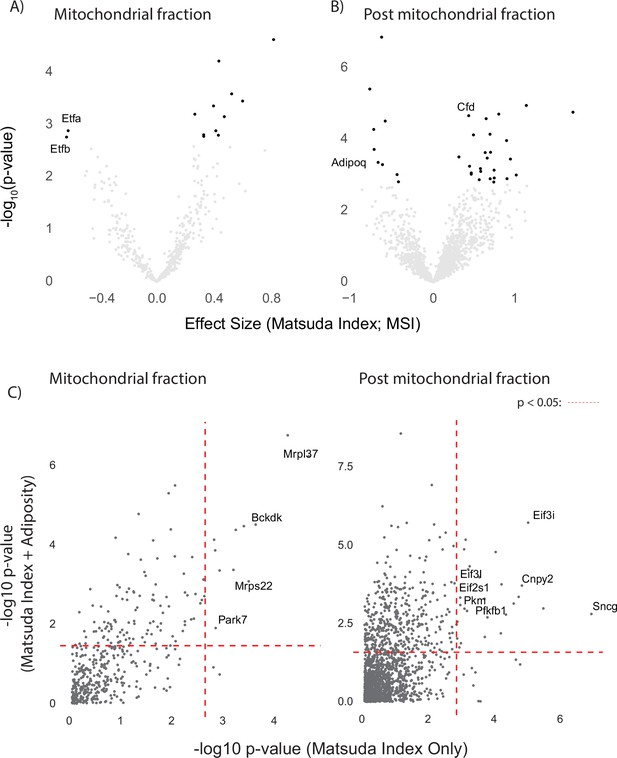
Comparison of linear modelling approaches for insulin sensitivity and muscle proteomics.
(A, B) Volcano plot with Matsuda Index effect sizes (x-axis) and significance (y-axis) for mitochondrial (A) and post-mitochondrial fraction (B) proteins. (C) Mitochondrial and post-mitochondrial fraction negative log10 p-values for Matsuda Index effect sizes from linear models with and without adiposity as a covariate. Dashed red line indicates false discovery rate-adjusted p-value<0.05.

Integration of proteomic data via Connectivity Map (CMAP).
(A, B) Workflow includes filtering for proteins with cis-pQTL (A) and negative association with Matsuda Index (B) prior to CMAP query. (A) Distribution of cis- and trans-pQTL across mitochondrial and post-mitochondrial fraction (PMF) proteome. (B) Left side of volcano plots from Figure 2A and B (proteins negatively associated with Matsuda Index) is shown with proteins comprised in molecular fingerprint of insulin resistance (IR) indicated in black. Proteins with human homologues are highlighted. (C) Comparison of molecular fingerprint of insulin resistance across muscle adipose and liver proteomes. (D) Top 10 KEGG pathways enriched in the molecular fingerprint of insulin resistance with proteins of interest highlighted. (E) Distribution of CMAP scores for identified small molecules and ligands with compounds of interest indicated. Significance testing was performed by chi-square test. * indicates a significant difference p<0.001. pQTL, protein quantitative trait loci.
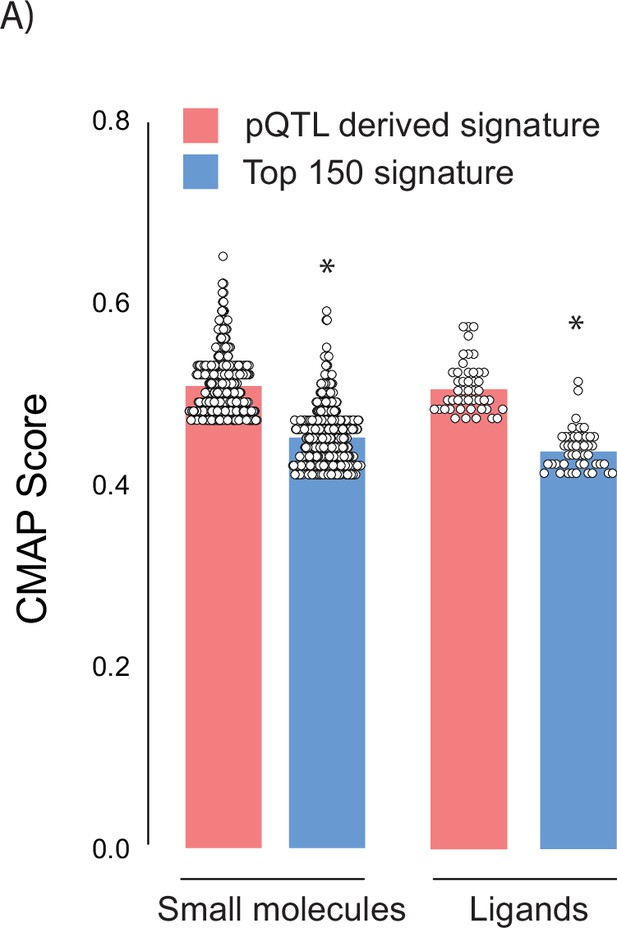
Assessment of protein quantitative trait loci (pQTL) filtration as a method to improve Connectivity Map (CMAP) compound identification.
(A) Raw CMAP scores for compounds and ligands identified by CMAP using either a pQTL filtered signature (fingerprint) or a signature of the 150 proteins with the largest negative effect sizes. Significance was determined by Student’s t-test. * indicates a significant difference between groups p<0.01.

Prestwick library of U.S. Food and Drug Administration (FDA)-approved drugs that modulate GLUT4 translocation in L6 myotubes.
(A) Schematic representation of the three assays performed. (B) Small molecules that promote GLUT4 exocytosis to the plasma membrane (PM) independently of insulin with controls (basal, insulin) on the left. (C) Small molecules that potentiate a submaximal dose of insulin (1 nM) with controls (basal, 1 nM and 100 nM insulin) on the left. Compounds were added in combination with 1 nM insulin. (D) Small molecules that reverse palmitate induced insulin resistance with controls (basal, insulin, insulin + palmitate) on the left. Compounds were added in combination with 100 nM insulin following palmitate treatment. (E) Venn diagram of compound overlap between assays. Biological significance for each assay was defined as 50% of corresponding control, see methods for details.
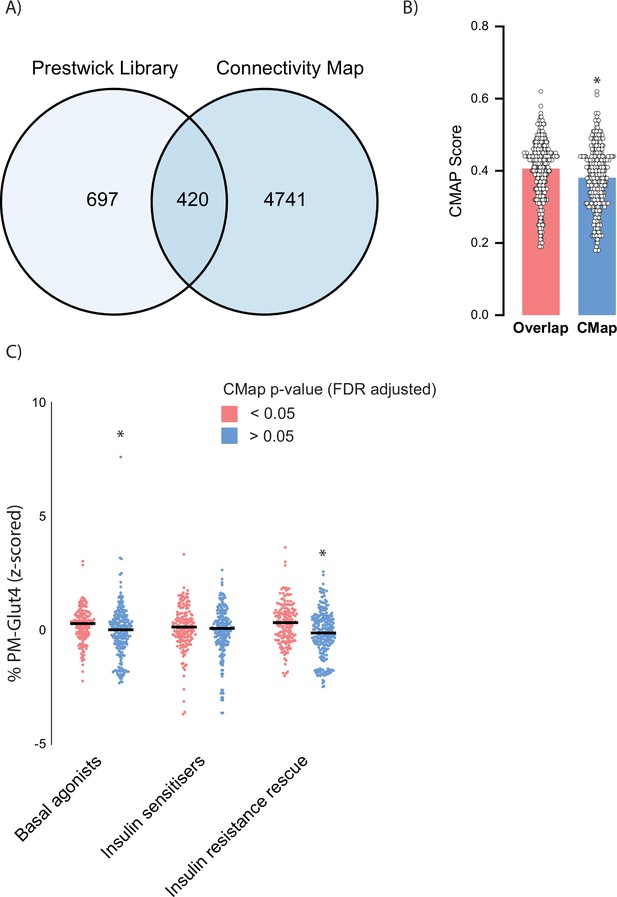
Validation of Connectivity Map (CMAP) results against Prestwick library of small molecules.
(A) Overlap of compounds found in both the Prestwick library and CMAP. (B) Comparison of CMAP scores between overlapping compounds and all of CMAP. (C) Comparison of z-scored GLUT4 at plasma membrane (PM) after treatment with Prestwick library compounds based on CMAP significance. Data are mean with individual data points. Significance was determined by two-way ANOVA with Student’s post hoc test or Student’s t-test. * indicates a significant difference between groups p<0.01.

Cross-validation of Connectivity Map (CMAP) and Prestwick library.
(A) Scoring matrix of top 20 scoring compounds present in both CMAP and the Prestwick library screens (basal agonists, Bas.Ag.; insulin sensitiser, Ins. Sens; insulin resistance reversers, Ins. Res. Rev). (B, C) Insulin stimulated GLUT4 translocation to the plasma membrane (PM) (B) and 2-deoxyglucose uptake (C) in control and insulin resistant L6 myotubes (palmitate) treated with thiostrepton or vehicle control. (D) Insulin-stimulated 2-deoxyglucose uptake in soleus and extensor digitorum longus (EDL) muscles from chow and Western diet (WD)-fed C57BL/6J and BXH9/TyJ following ex vivo treatment with thiostrepton or vehicle control. Data are mean with individual data points shown, n = 3–4 (B, C), n = 3–5 (BXH9); n = 11–14 (C57BL/6J). Significance was determined by one-way ANOVA with Student’s post hoc test. ** indicates significant difference from control (basal or chow-fed C57BL6/J) group (p<0.01), * indicates significant difference from control (p<0.05). # indicates significant difference from palmitate-treated or WD-fed control group (p<0.05).
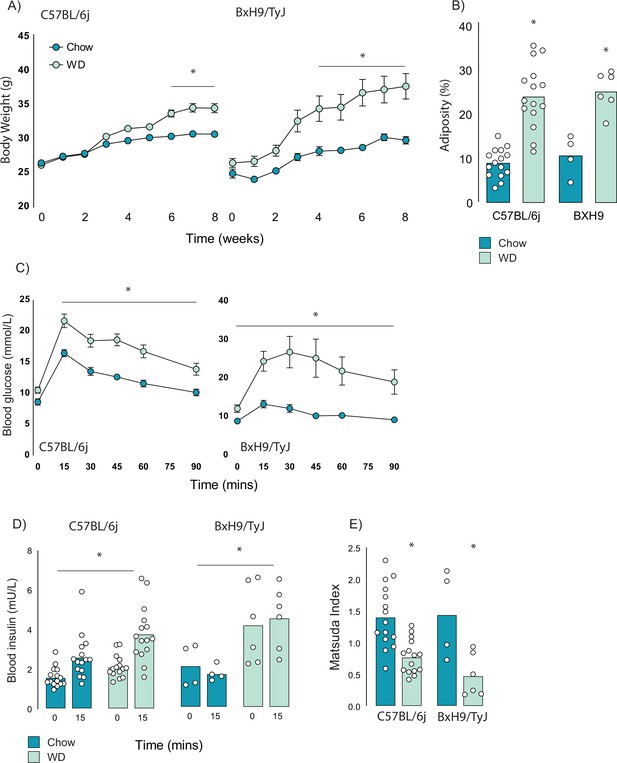
Effect of Western diet (WD) feeding on C57BL/6J and BxH9/TyJ mice body composition and insulin sensitivity.
(A–E) Body weights (A), adiposity (B), blood glucose (C), and insulin (D) during a glucose tolerance test, and Matsuda Index (E) of C57BL/6J and BxH9/TyJ mice fed a chow of high-fat/high-sugar (WD) for 8 wk. Data are mean with individual data points, n = 4–6 (BxH9), n = 15 (C57BL/6J).
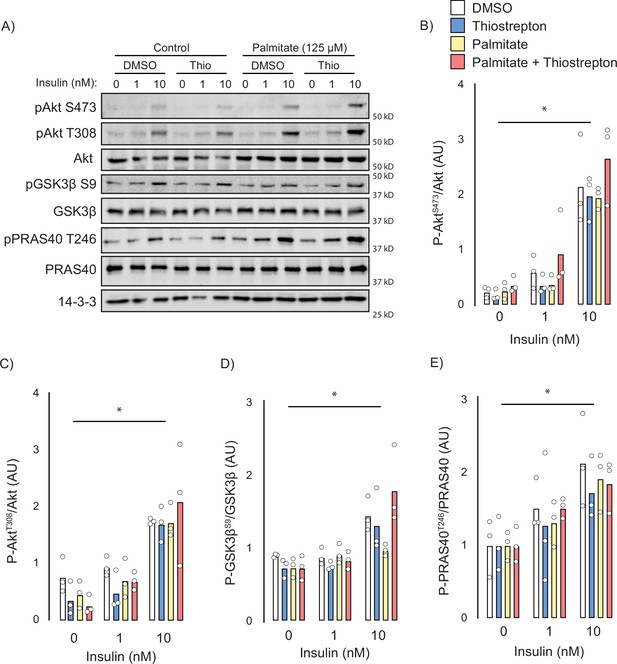
Effect of thiostrepton on insulin signalling.
(A–E) Immunoblotting with indicated antibodies of control and palmitate treated GLUT4-HA-L6 myotubes following treatment with thiostrepton or vehicle control and stimulation with 0, 1, or 10 nM insulin. (A) Representative immunoblot shown of three independent experiments. (B–E) Quantification of immunoblots in (A) Akt S473 (B), Akt T308 (C), GSK3β S9 (D), and PRAS40 T246 (E). Data are mean with individual data points, n = 3. Significance was determined by two-way ANOVA with Student’s post hoc test. * indicates a significant effect of insulin p<0.01.
-
Figure 6—source data 1
Uncropped immunoblots with indicated antibodies of control and palmitate treated GLUT4-HA-L6 myotubes following treatment with thiostrepton or vehicle control and stimulation with 0, 1, or 10 nM insulin.
Representative immunoblot shown of three independent experiments.
- https://cdn.elifesciences.org/articles/86961/elife-86961-fig6-data1-v1.zip
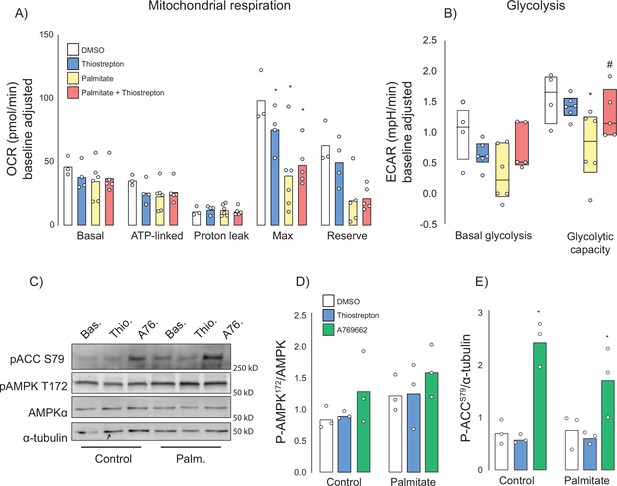
Effect of thiostrepton on mitochondrial respiration and glycolysis.
(A, B) Oxygen consumption rates (A) and extracellular acidification rates (B) in control and palmitate treated GLUT4-HA-L6 myotubes treated with either thiostrepton or vehicle control. (C) Immunoblotting of AMPK signalling in control and palmitate-treated (Palm.) GLUT4-HA-L6 myotubes treated with either thiostrepton (Thio.), vehicle control (Bas.) or positive control A-769662 (A76). Representative immunoblots shown of three independent experiments. (D, E) Quantification of immunoblots in (C) AMPK T172 (D) and ACC S79 (E) phosphorylation following thiostrepton or A-769662 treatment. Data are mean with individual data points shown, n = 3. * indicates significant difference from control cells, # indicates significant difference from palmitate-treated cells p<0.05.
Additional files
-
Supplementary file 1
List of proteins and their Matsuda Index effect sizes which comprise our pQTL-filtered molecular fingerprint of insulin resistance.
- https://cdn.elifesciences.org/articles/86961/elife-86961-supp1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/86961/elife-86961-mdarchecklist1-v1.docx





