Tetraose steroidal glycoalkaloids from potato provide resistance against Alternaria solani and Colorado potato beetle
Figures
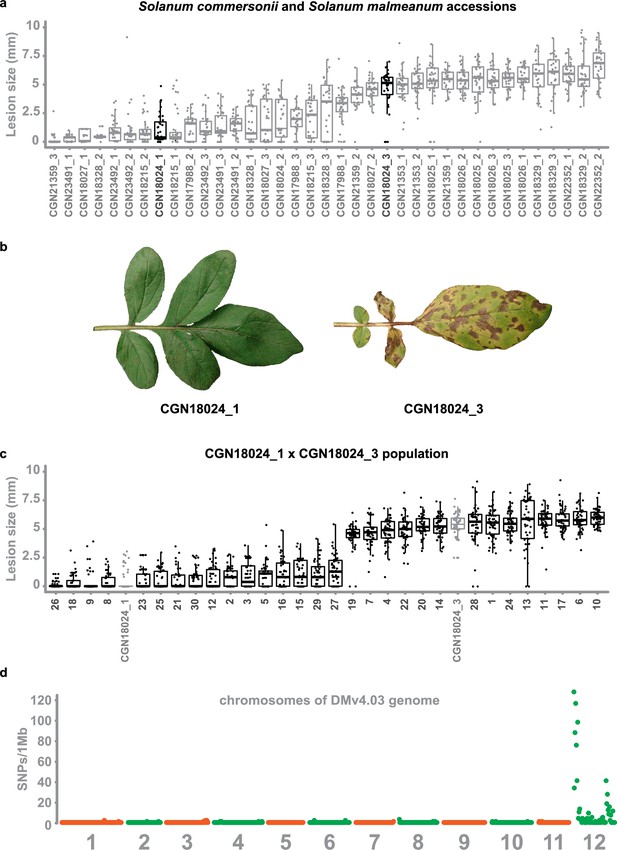
Early blight resistance maps to chromosome 12 of potato.
(a) Two to three genotypes of 13 different accessions of S. commersonii and S. malmeanum were inoculated with A. solani altNL03003. Three plants of each genotype were tested and three leaves per plants were inoculated with six 10 µl droplets with spore suspension. Lesion diameters were measured 5 days post inoculation and visualised using boxplots, with horizonal lines indicating median values and individual measurements plotted on top. Non-expanding lesions (<3 mm) indicate resistance and expanding lesions indicate susceptibility. Some accessions segregate for resistance. (b) Accession CGN18024 is an example of an accession that segregates for resistance to A. solani, with CGN18024_1 displaying resistance and CGN18024_3 displaying susceptibility at 5 days after spray-inoculation. (c) Progeny from CGN18024_1 × CGN18024_3 was inoculated with A. solani. Three plants of each genotype were tested and three leaves per plants were inoculated with six 10 µl droplets with spore suspension each. Lesion diameters were measured 5 days post inoculation. 16 progeny genotypes are resistant (with lesion diameters <2–3 mm) and 14 are susceptible (with expanding lesions). This corresponds to a 1:1 segregation ratio (Χ2 (1, N = 30) = 0.133, ρ = 0.72). (d) SNPs derived from a BSR-Seq analysis (heterozygous in resistant parent CGN18024_1 and the resistant bulk, but absent or homozygous in susceptible parent and susceptible bulk) using bulks of susceptible and resistant progeny were plotted in 1 Mb windows over the 12 chromosomes of the potato DMv4.03 genome (Xu et al., 2011). They are almost exclusively located on chromosome 12.
-
Figure 1—source data 1
Numerical data underlying Figure 1a.
- https://cdn.elifesciences.org/articles/87135/elife-87135-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Numerical data underlying Figure 1c.
- https://cdn.elifesciences.org/articles/87135/elife-87135-fig1-data2-v1.xlsx
-
Figure 1—source data 3
Numerical data underlying Figure 1d.
- https://cdn.elifesciences.org/articles/87135/elife-87135-fig1-data3-v1.xlsx
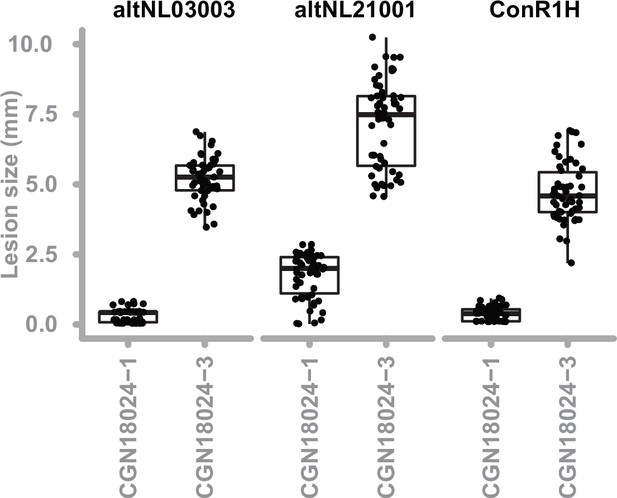
Resistance of S commersonii genotypes CGN18024_1 and CGN18024_3 against different isolates of A.solani.
CGN18024_1 and CGN18024_3 (3 replicates per genotypes, three leaves per plant and six inoculations per leaf) were inoculated with A. solani altNL03003 (the reference isolate used throughout the manuscript), altNL21001 (isolated from a potato field in the Netherlands in 2021), and ConR1H (harvested from a potato field in Idaho, USA in 2015). CGN18024_1 is resistant against all three isolates, whereas CGN18024_3 is susceptible.
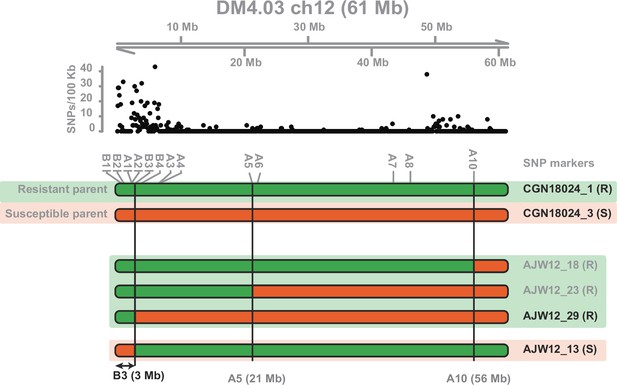
Resistance fromS.commersonii to A. solani is mapped to the top of chromosome 12.
Filtered SNPs from bulked segregant RNAseq analysis (BSA-RNAseq) are plotted in 100 kb windows on chromosome 12 of the DMv4.03 genome at the top of the figure. A selection of SNPs (‘A1’–‘A10’ and ‘B1’–‘B4’) was used as markers in high-resolution melting (HRM) analysis to genotype resistant S commersonii parent CGN18024_1 and susceptible parent CGN18024_3 from the AJW12 mapping population as well as progeny used in BSA-RNAseq. HRM analysis led to the identification of recombinants AJW12_13, AJW12_18, AJW12_23, and AJW12_29. Recombinant AJW12_13 (susceptible to A. solani) and recombinant AJW12_29 (resistant to A. solani) are used to map the resistance locus from S. commersonii to a window of approximately 3 Mb at the top of chromosome 12, delimited by marker ‘B3’.
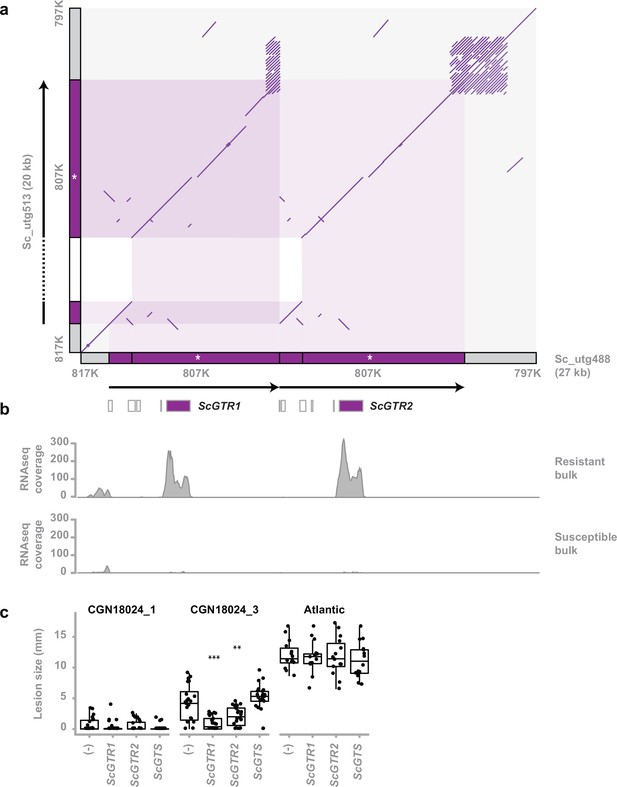
Identification of two glycosyltransferase resistance genes.
(a) Comparison of the susceptible and resistant haplotype of the Solanum commersonii CGN18024_1 resistance region (delimited by markers 817K and 797K) in a comparative dot plot shows a rearrangement. Locations of markers used to map the resistance region are indicated in grey along the x- and y-axis. The duplicated region of the resistant haplotype contains marker 807K (white asterisk) and two predicted glycosyltransferase genes (ScGTR1 and ScGTR2). Several short ORFs with homology to glycosyltransferase genes that were predicted in the resistance region are indicated by white boxes, but ScGTR1 and ScGTR2 are the only full-length genes. As a result of the rearrangement, the resistance region of the resistant haplotype (27 kb) is 7 kb larger than the corresponding region of the susceptible haplotype (20 kb). (b) Alignment of RNAseq reads from the BSR-Seq analysis shows that ScGTR1 and ScGTR2 are expressed in bulks of resistant progeny, but not in bulks of susceptible progeny. (c) S. tuberosum cv. ‘Atlantic’, S. commersonii CGN18024_1 and CGN18024_3 were agroinfiltrated with expression constructs for ScGTR1 and ScGTR2, ScGTS, and empty vector (-), combined as seperate spots on the three middle leaves of each genotype (8 plants per genotype). A. solani is inoculated in the middle of agroinfiltrated areas at 2 days after agroinfiltration and lesion diameters are measured 5 days after inoculation. Lesion sizes were visualised with boxplots, with horizonal lines indicating median values and individual measurements plotted on top. Agroinfiltration with expression constructs for ScGTR1 and ScGTR2 results in a significant (Welch’s two-sample t-test, **p < 0.01, ***p < 0.001) reduction of lesion sizes produced by Alternaria solani altNL03003 in S. commersonii CGN18024_3, but not in S. tuberosum cv. ‘Atlantic’.
-
Figure 2—source data 1
Numerical data underlying Figure 2c.
- https://cdn.elifesciences.org/articles/87135/elife-87135-fig2-data1-v1.xlsx
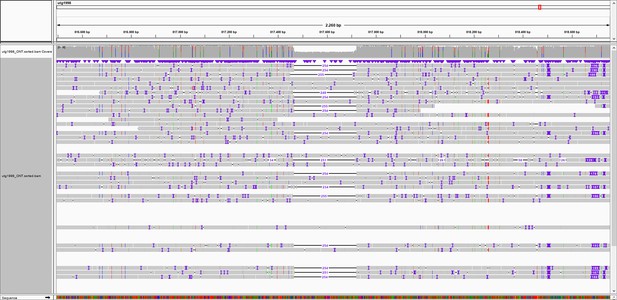
Overview of marker 817K.
Integrated Genomics Viewer (IGV) snapshot of Oxford Nanopore Technology (ONT) reads aligned to the genome of S. commersonii CGN18024_1. An Insertion/Deletion (InDel) of 254 bp is observed at approximately 817 kb of contig utg1998 that covers the resistance region. Primers were designed flanking the InDel to develop marker 817K.
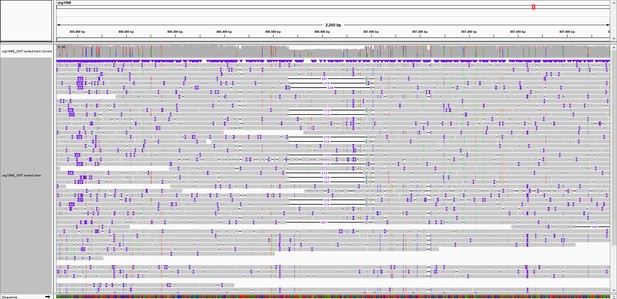
Overview of marker 807K.
Integrated Genomics Viewer (IGV) snapshot of Oxford Nanopore Technology (ONT) reads aligned to the genome of S. commersonii CGN18024_1. An Insertion/Deletion (InDel) of 310 bp is observed at approximately 807 kb of contig utg1998 that covers the resistance region. Primers were designed flanking the InDel to develop marker 807K.
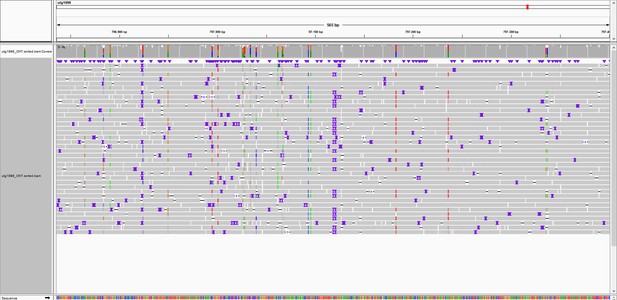
Overview of marker 797K.
Integrated Genomics Viewer (IGV) snapshot of Oxford Nanopore Technology (ONT) reads aligned to the genome of S. commersonii CGN18024_1. An Insertion/Deletion (InDel) of 6 bp is observed at approximately 797 kb of contig utg1998 that covers the resistance region. Primers were designed flanking the InDel to develop marker 797K.
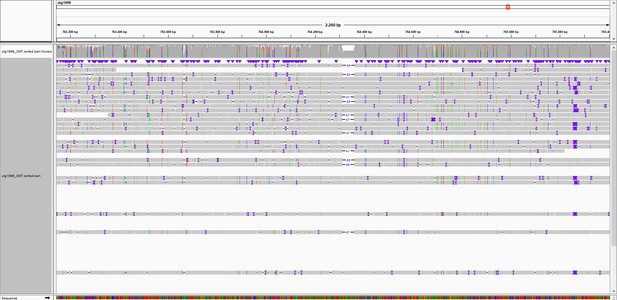
Overview of marker 764K.
Integrated Genomics Viewer (IGV) snapshot of Oxford Nanopore Technology (ONT) reads aligned to the genome of S. commersonii CGN18024_1. An Insertion/Deletion (InDel) of 47 bp is observed at approximately 764 kb of contig utg1998 that covers the resistance region. Primers were designed flanking the InDel to develop marker 764K.
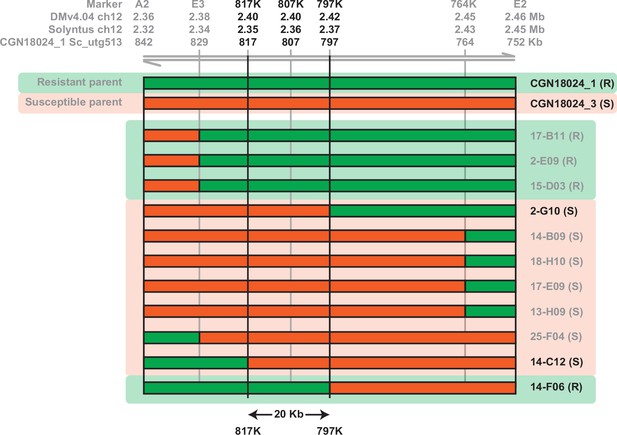
Fine mapping the resistance locus in CGN18024_1.
New markers based on the Solyntus and CGN18024_1 genome were used to screen for recombinants among progeny from a cross between resistant CGN18024_1 and susceptible CGN18024_3. Physical locations of the markers on the DMv4.04, Solyntus, and CGN18024_1 genome are indicated at the top of the figure. Recombinants that were identified were tested for resistance to A. solani to fine map the resistance region. Recombinants 2-G10 (resistant, R), 14-F06 and 14-C12 (both susceptible, S) are used to delimit the resistance region between markers 817K and 797K, corresponding to a region of 20 kb in the CGN18024_1 genome.
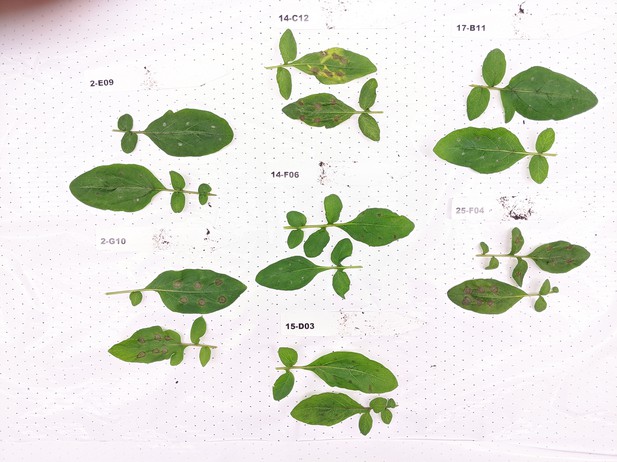
Early blight disease symptoms on key recombinants.
The picture shows lesions of representative leaves of key recombinants at 5 days post drop inoculation with spores of A. solani.

Alignment of putative S. commersonii glycosyltransferases (ScGTs) linked to resistance.
ScGTR1, ScGTR2, and ScGTS show high similarity, but the GT encoded by the susceptible haplotype (ScGTS) contains a mutation that leads to a truncated protein (stop codons are indicated with *).

Comparative phylogenetic analysis of glycosyltransferases with a known function (Supplementary file 2).
The phylogenetic tree is constructed using the maximum likelihood method (100 bootstraps). ScGTR1 and ScGTR2 are indicated with arrows and GTs with a previously characterised role in steroidal glycoalkaloid (SGA) biosynthesis are marked with *. Direct homologs of these SGA GTs (based on identity and synteny) derived from the CGN18024_1 genome are included in the analysis (names starting with ‘SCM’).

Leaf compounds from resistant S. commersonii inhibit growth of diverse fungi, including pathogens of potato.
(a) Crude leaf extract from CGN18024_1/CGN18024_3 was added to PDA plates (5%, wt/vol) and autoclaved (left) or semi-sterilised for 15 min at 60°C (right). Growth of Alternaria solani altNL03003 was strongly inhibited on PDA plates with autoclaved leaf extract from CGN18024_1 compared to plates with CGN18024_3, as shown on the left two pictures taken at 7 days after placing an agar plug with mycelium of A. solani at the centre of each plate. Abundant fungal contamination appeared after 4 days on plates containing semi-sterilised leaf from CGN18024_3, but not on plates containing material from CGN18024_1 (right two pictures). (b) Growth of potato pathogenic fungi A. solani (altNL03003), B. cinerea (B05.10), and F. solani (1992 vr) was followed by measuring the colony diameter on PDA plates containing autoclaved leaf material from CGN18024_1/CGN18024_3. Growth of all three fungi was measured on PDA plates containing CGN18024_1 (red squares), CGN18024_3 (green circles), or plates with PDA and no leaf material (blue triangles). Three replicates were used per isolate/substrate combination. Significant differences in growth on PDA plates containing plant extract compared to PDA plates without leaf extract are indicated with asterisks (Welch’s two-sample t-test, **p < 0.01, ***p < 0.001).
-
Figure 3—source data 1
Numerical data underlying Figure 3b.
- https://cdn.elifesciences.org/articles/87135/elife-87135-fig3-data1-v1.xlsx

Tetraose steroidal glycoalkaloids (SGAs) from Solanum commersonii provide resistance to Alternaria solani and Colorado potato beetle.
Data are visualised with boxplots, with horizonal lines indicating median values and individual measurements plotted on top. (a) Tetraose SGAs were detected in resistant CGN18024_1 and in CGN18024_3 transformed with ScGTR1/ScGTR2. Susceptible S. tuberosum cv. ‘Atlantic’ and wildtype (WT) CGN18024_3 contain only triose SGAs. Overexpression of ScGTR1 resulted in the addition of a hexose to the triose SGAs from CGN18024_3, resulting in a commertetraose (Gal-Glu-Glu-Glu), while overexpression of ScGTR2 caused the addition of a pentose, resulting in a lycotetraose (Gal-Glu-Glu-Xyl). Each boxplot displays the data of three seperate measurements (b) WT CGN18024_1/CGN18024_3 and CGN18024_3 transformants were inoculated with Alternaria solani altNL03003. Three plants of each genotype were tested and three leaves per plants were inoculated with six 10 µl droplets with spore suspension each. Lesions diameters were measured 5 days post inoculation. Significant differences with WT CGN18024_3 are indicated with asterisks (Welch’s two-sample t-test, ***p < 0.001). ScGTR1 and ScGTR2 can both complement resistance to A. solani in CGN18024_3, as the lesion sizes produced on CGN18024_3 transformants are comparable to resistant CGN18024_1. (c) Three plants per genotype were challenged with five Colorado potato beetle larvae each. The tetraose SGAs produced by ScGTR1 and ScGTR2 can provide resistance to Colorado potato beetle, as indicated by reduced larvae survival and total larvae weight. Significant differences with WT CGN18024_3 are indicated with asterisks (Welch’s two-sample t-test, *p < 0.05, ***p < 0.001). (d) Putative structures of SGAs detected in CGN18024_1 and CGN18024_3, based on previous studies (Osman et al., 1976; Distl and Wink, 2009; Caruso et al., 2011; Vázquez et al., 1997). CGN18024_3 produces triose SGAs and is susceptible to Colorado potato beetle and A. solani. ScGTR1 and ScGTR2 from CGN18024_1 convert these triose SGAs from susceptible S. commersonii to tetraose SGAs, through the addition of a glucose or xylose moiety, respectively. Both sugar additions can provide resistance to Colorado potato beetle and A. solani.
-
Figure 4—source data 1
Numerical data underlying Figure 4a.
- https://cdn.elifesciences.org/articles/87135/elife-87135-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Numerical data underlying Figure 4b.
- https://cdn.elifesciences.org/articles/87135/elife-87135-fig4-data2-v1.xlsx
-
Figure 4—source data 3
Numerical data underlying Figure 4c.
- https://cdn.elifesciences.org/articles/87135/elife-87135-fig4-data3-v1.xlsx
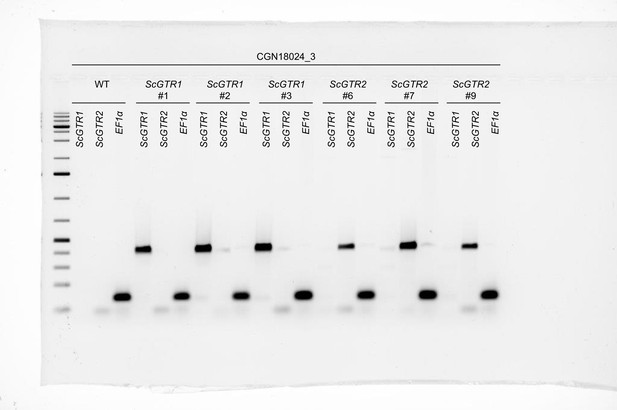
Validation of ScGTR1 and ScGTR2 transformants using PCR.
Gel electrophoresis of PCR amplicons produced by primer combinations p35S + ScGTR1sr3 (ScGTR1), p35S + ScGTR2sr3 (ScGTR2) and ef1αF1 + ef1αR1 (EF1α) using genomic DNA template of wildtype CGN18024_3 (WT) and ScGTR1/ScGTR2 CGN18024_3 transformants. 6 µl of Kb Plus DNA Ladder is loaded in the first lane (with the lowest band corresponding to a DNA fragment of 75 bp and the first thick band from the bottom corresponding to a DNA fragment of 500 bp).
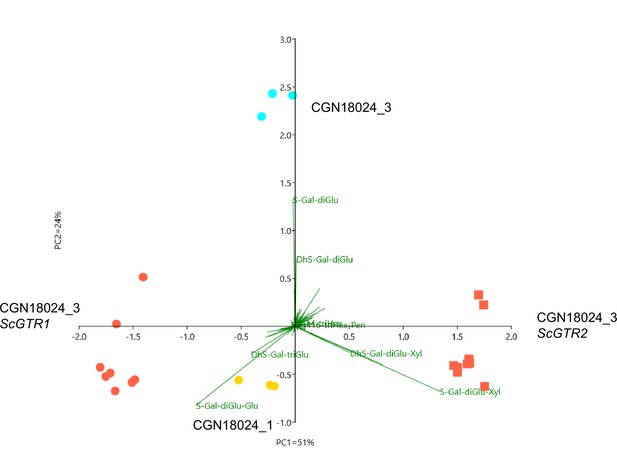
Principal component analysis (PCA) on Solanum commersonii genotypes and transformants.
PCA based on 1041 mass peaks detected by ultra high-performance liquid chromatography (UPLC)–mass spectrometry (MS) in leaves of ScGTR1 (red dots) and ScGTR2 (red squares) transformants compared to the corresponding susceptible wildtype Solanum commersonii CGN18024_3 (blue circles) and resistant CGN18024_1 (yellow circles). Three independent transformants and three replicates per genotype were analysed. 75% of the total metabolic variation between the groups is explained by the first and the second PC, mostly loaded by variation between tri- and tetraglycosylated steroidal glycoalkaloids. S/DhS – solanidine/demissidine.
Tables
Genome assembly metrics of S. commersonii cmm1t (Aversano et al., 2015) and CGN18024_1.
| Genome | CMM1t* | CGN18024_1 |
|---|---|---|
| Total size (Mb) | 730 | 737 |
| Contig number | 278,460 | 637 |
| Largest contig (Mb) | 0.17 | 21.2 |
| N50 (Mb) | 0.007 | 4.02 |
| Complete BUSCO (%) | 81.9 | 95.7 |
-
*
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Solanum commersonii) | ScGTR1 | This paper, sequence deposited at GenBank | GenBank: OM830430 | |
| Gene (S. commersonii) | ScGTR2 | This paper, sequence deposited at GenBank | GenBank: OM830431 | |
| Strain, strain background (S. commersonii) | CGN18024 | CGN WUR | CGN18024 | |
| Strain, strain background (Alternaria solani) | altNL03003 | Wolters et al., 2019; CBS-KNAW | altNL03003; CBS 143772 | |
| Genetic reagent (S. commersonii) | CGN18024_3-ScGTR1-1,2 and 3 | This paper | Maintained at plant breeding, WUR; available upon reasonable request | |
| Genetic reagent (S. commersonii) | CGN18024_3-ScGTR2-6,7 and 9 | This paper | Maintained at plant breeding, WUR; available upon reasonable request |
Additional files
-
Supplementary file 1
Solanum commersonii and Solanum malmeanum accessions used in this study.
Accessions were obtained from the Centre for Genetic Resources, the Netherlands (CGN WUR). Two to three genotypes from each accession were used in the disease screen with A. solani.
- https://cdn.elifesciences.org/articles/87135/elife-87135-supp1-v1.xlsx
-
Supplementary file 2
Overview of characterised glycosyltransferases (GTs) used in comparative phylogenetic analysis (Figure 2—figure supplement 8).
GTs with a known function are taken from Bowles et al., 2005, McCue et al., 2005, McCue et al., 2006, McCue et al., 2007 , Masada et al., 2009, Itkin et al., 2013, Itkin et al., 2011, and Tikunov et al., 2013.
- https://cdn.elifesciences.org/articles/87135/elife-87135-supp2-v1.xlsx
-
Supplementary file 3
Putative identities and relative contents of steroidal glycoalkaloids (SGAs) in different potato genotypes.
Average signal intensities (3 replicates per genotype) are presented as a percentage of the maximum signal intensity.
- https://cdn.elifesciences.org/articles/87135/elife-87135-supp3-v1.xlsx
-
Supplementary file 4
Overview of the steroidal glycoalkaloids detected in our study.
RT – retention time; [M−H+FA]− – mass of a molecular ion at negative ionisation mode (all alkaloids were represented by formic acid adduct ions); [M+H]+ – mass of a molecular ion at negative ionisation mode; Putative structure – putative combination of aglycones and sugar moieties deduced by comparing the fragmentation spectrum derived at positive ionisation with previous studies (Osman et al., 1976; Distl and Wink, 2009; Caruso et al., 2011; Vázquez et al., 1997); Fragmentation spectra derived using positive ionisation: P – parent ion or P-fragment(s) loss.
- https://cdn.elifesciences.org/articles/87135/elife-87135-supp4-v1.xlsx
-
Supplementary file 5
Primers used to map the resistance region.
- https://cdn.elifesciences.org/articles/87135/elife-87135-supp5-v1.xlsx
-
Supplementary file 6
Primers used to clone candidate resistance genes.
- https://cdn.elifesciences.org/articles/87135/elife-87135-supp6-v1.xlsx
-
Supplementary file 7
Primers used to validate transformants.
- https://cdn.elifesciences.org/articles/87135/elife-87135-supp7-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/87135/elife-87135-mdarchecklist1-v1.docx





