A local ATR-dependent checkpoint pathway is activated by a site-specific replication fork block in human cells
Figures
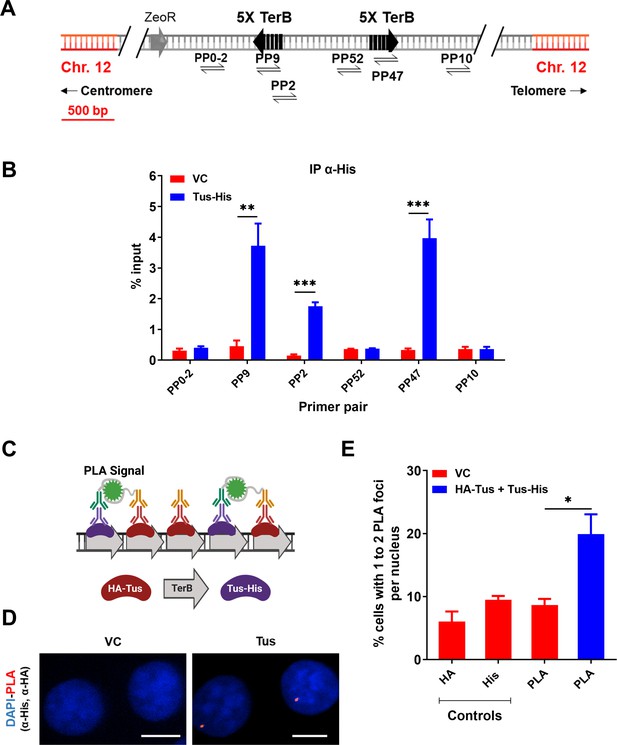
Tus is bound to TerB sites integrated in human cells.
(A) Schematic of linearized plasmid (pWB15) integrated into MCF7 cells (MCF7 5C-TerB) with two cassettes containing five tandem TerB sequences in the non-permissive orientation (black arrows). Black half-arrowheads depict polymerase chain reaction (PCR) products expected from stated primer pairs used in (B). (B) Chromatin immunoprecipitation (ChIP) with His antibody on MCF7 5C-TerB cells transfected with either a vector control (VC) or HA-Tus-His expression plasmids. ChIP-qPCR was conducted using the indicated primer pairs. Data show the percentage of input (n = 3). (C) Schematic of proximity ligation assay (PLA) to visualize Tus bound to TerB sites using HA and His antibodies. (D) Representative images of the PLA foci across stated conditions. Scale Bars, 10 μm (E) Percentage of cells with 1–2 PLA foci per nucleus. (n = 3, ≥150 cells per experiment). Bar graphs represents mean ± standard error of the mean (SEM), Student t-test (see methods).
-
Figure 1—source data 1
Tables related to Figure 1B and E.
- https://cdn.elifesciences.org/articles/87357/elife-87357-fig1-data1-v1.zip
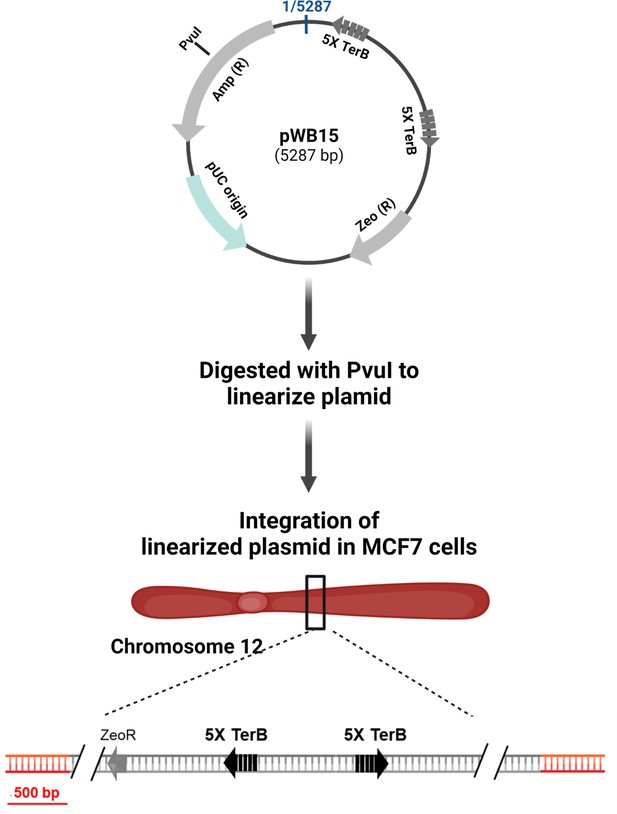
Generation of the MCF7 5C-TerB cell line.
Schematic of the integration of pWB15 in MCF7 cells to generate MCF7 5C-TerB. pWB15 contains two TerB cassettes (gray arrows). Each TerB cassette contains five tandem TerB sequences, which are in the non-permissive orientation in pWB15 (gray arrow facing away from each other). The plasmid was linearized by digesting with PvuI for integration into MCF7 cells. The single-copy integrant was confirmed using whole-genome sequencing and found to be integrated at chromosome 12.
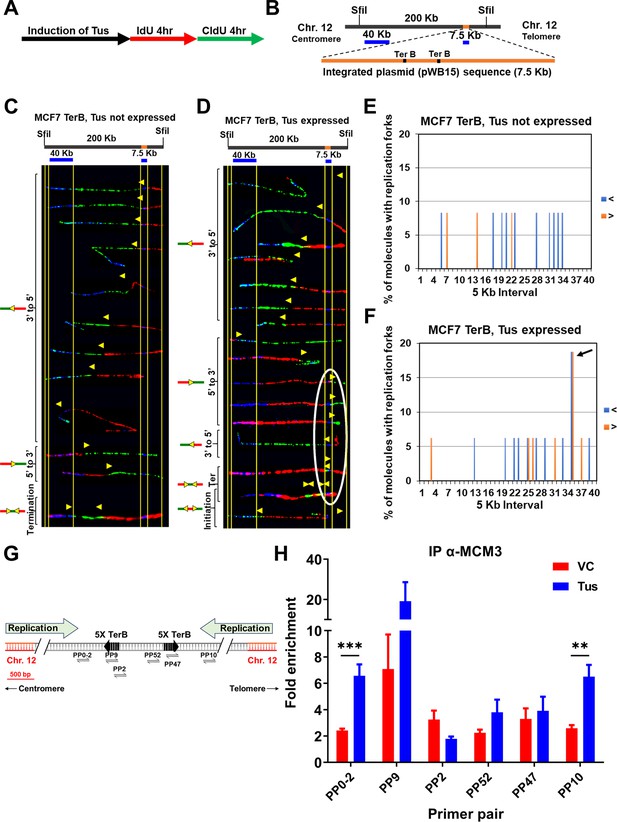
Replication fork pauses at the Ter sequence in the presence of the Tus protein.
(A) Schematic of pulse labeling of MCF7 5C-TerB cells with Tus protein expression for 3 d. (B) Locus map of a 200 kb SfiI segment from MCF7 5C-TerB Chromosome 12 with the integrated plasmid (pWB15) DNA (orange). A 40 kb FISH probe made from fosmid WI2-1478M20 and a 7.5 kb FISH probe made from plasmid pWB15 are shown in blue. (C, D) Top: locus map of the DNA segment containing Ter sequence with the location of the FISH probes. Bottom: photomicrographs of labeled DNA molecules from MCF7 5C-TerB, Tus not expressed (C) and MCF7 5C-TerB, Tus expressed (D). Yellow arrows indicate the position of replication forks at the transition of labeling from IdU to CldU incorporation showing replication fork direction. Molecules are arranged in the following order: forks progressing from 3′ to 5′, forks progressing from 5′ to 3′, termination events, and initiation events. Replication forks (yellow arrows) in the white oval are all at the same location (at the TerB sequence) on molecules from different cells indicating that replication forks are pausing at this TerB sequence. (E, F) Percentage of molecules with replication forks at each 5 kb interval in the 200 Kb SfiI segment containing TerB sequence, quantified from molecules in MCF7 5C-TerB (E) Tus not expressed and (F) Tus expressed. Percentage of molecules with replication forks progressing 3′ to 5′ (< blue) and 5′ to 3′ (> orange) are shown. In the cells expressing Tus, a high percentage of molecules contain replication forks pausing in both directions in the 5 kb interval that contains Ter sequences (black arrow [F], white oval [D]). (G) Schematic similar to Figure 1A with progression of the endogenous origins of replication within the chromosome 12 shown (green arrows). (H) Chromatin immunoprecipitation (ChIP) using MCM3 antibody on MCF7 5C-TerB cells transfected with vector control (VC) or HA-Tus-His plasmids. ChIP-qPCR was conducted using the indicated primer pairs. Data shows the fold enrichment relative to the IgG controls (n = 3). Bar graphs represents mean ± standard error of the mean (SEM), Student t-test (see methods).
-
Figure 2—source data 1
Table related to Figure 2H.
- https://cdn.elifesciences.org/articles/87357/elife-87357-fig2-data1-v1.zip
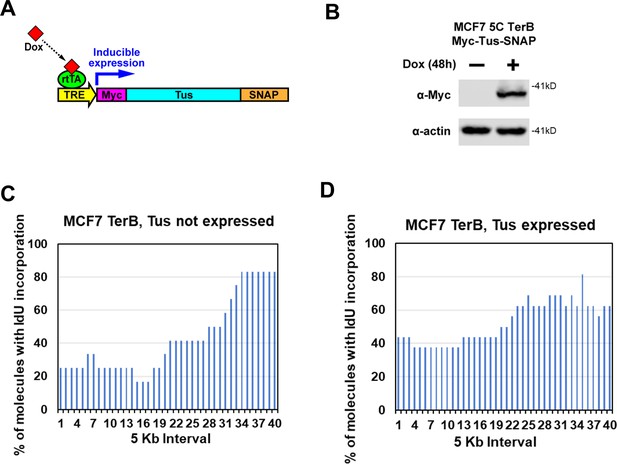
Expression of Tus in MCF7 5C-TerB cells.
(A) Schematic of doxycycline (Dox)-inducible expression of Myc-NLS-TUS-SNAP. (rtTA = reverse tetracycline trans activator; TRE = tetracycline response element). (B) Immunoblot of MCF7 5C-TerB cells stably expressing Dox-inducible Myc-NLS-TUS-SNAP. (C, D) Replication profiles shown as the percentage of molecules with IdU incorporation at each 5 kb interval in the 200 kb SfiI segment containing TerB sequence, quantified from molecules MCF7 5C-TerB in Figure 2C (Tus not expressed) and Figure 2D (Tus expressed).
-
Figure 2—figure supplement 1—source data 1
Unedited western blot images for Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/87357/elife-87357-fig2-figsupp1-data1-v1.zip
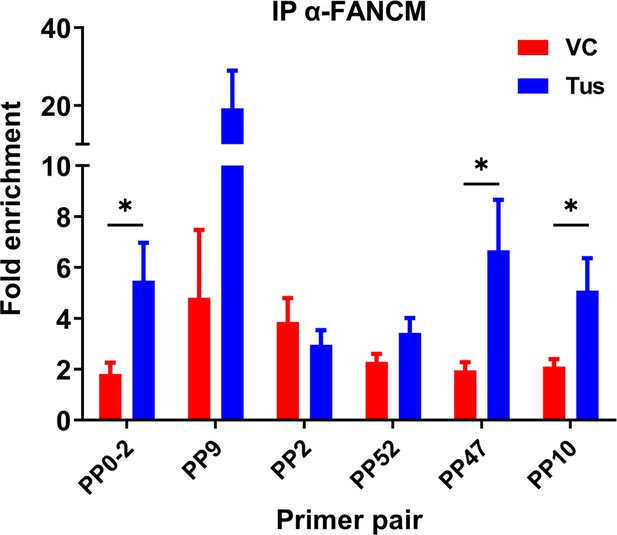
Enrichment of FANCM at the Ter sequence in the presence of the Tus protein.
Chromatin immunoprecipitation (ChIP) using FANCM antibody was performed MCF7 5C-TerB cells transfected with vector control (VC) or HA-Tus-His plasmids. ChIP-qPCR was conducted using the indicated primer pairs. Data shows the fold enrichment relative to the IgG controls (n = 3). Bar graphs represents mean ± standard error of the mean (SEM), Student t-test (see methods).
-
Figure 2—figure supplement 2—source data 1
Table related Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/87357/elife-87357-fig2-figsupp2-data1-v1.zip
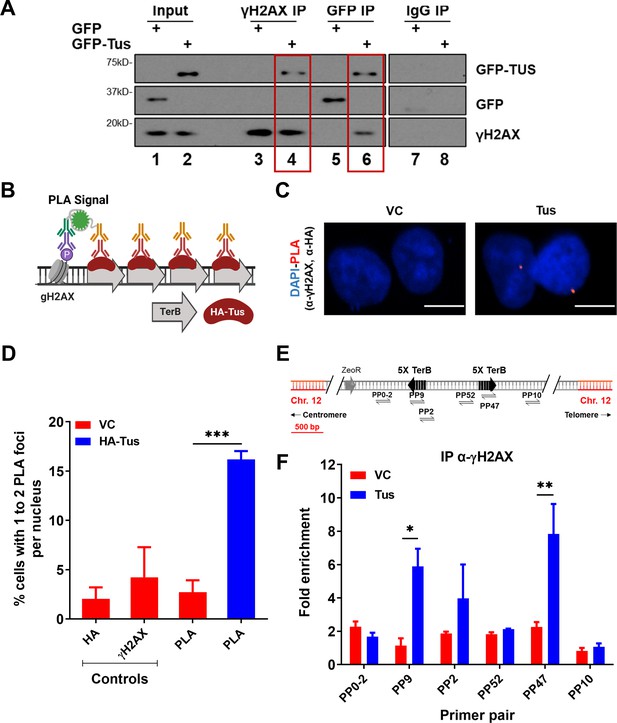
γH2AX is enriched at TerB sites after Tus expression.
(A) MCF7 5C-TerB cells transfected with GFP or GFP-Tus, γH2AX and IgG antibodies were used to immuno-precipitate proteins and analyzed by immunoblotting with indicated antibodies. (B) Schematic of proximity ligation assay (PLA) to visualize HA-Tus bound to TerB in the proximity of γH2AX sites using HA and γH2AX antibodies. (C) Representative images of the PLA foci across stated conditions. Scale Bars, 10 μm. (D) Percentage of cells with 1–2 PLA foci per nucleus (n = 3, ≥150 cells per experiment). (E) Schematic of MCF7 5C-TerB depicting the positions of the primer pairs with respect to the integrated TerB locus. (F) γH2AX levels along the integrated TerB plasmid were analyzed by ChIP-qPCR in MCF7 5C-TerB cells transfected with vector control (VC) or Tus expression plasmids using the indicated primer pairs. Data shows the fold enrichment relative to IgG controls (n = 3). Bar graphs represents mean ± standard error of the mean (SEM), Student t-test (see methods).
-
Figure 3—source data 1
Unedited western blot images for Figure 3A.
- https://cdn.elifesciences.org/articles/87357/elife-87357-fig3-data1-v1.zip
-
Figure 3—source data 2
Tables related to Figure 3D and F.
- https://cdn.elifesciences.org/articles/87357/elife-87357-fig3-data2-v1.zip
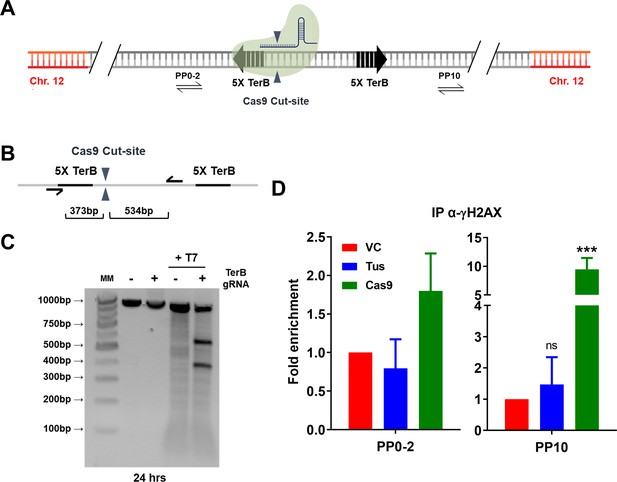
Distinct patterns of γH2AX enrichment at a Cas9-mediated DSB versus the Tus-TerB fork barrier.
(A) Schematic of the linearized TerB plasmid (pWB15) integrated as a unique copy into MCF7 cells (MCF7 5C-TerB) with the Cas9-binding site (green protein). Blue triangles: Cas9 cut site. Black half-arrowheads depict PCR products expected from primer pair (PP10) used in quantitative PCR (qPCR) are shown. (B) Schematic depicting the Cas9 cut site (blue arrows), site-specific PCR primers (black half-arrowhead), and predicted size of cleavage products. (C) PCR analysis by T7 assay of MCF7 5C-TerB cells transfected with Cas9-sgRNA RNP complex. (D) γH2AX levels along the integrated TerB plasmid were analyzed by ChIP-qPCR in MCF7 5C-TerB cells transfected with vector control (VC), Tus, or Cas9 expression plasmids using the indicated primer pairs. Data shows the fold enrichment relative to IgG controls (n = 2). Bar graphs represents mean ± standard error of the mean (SEM), Student t-test (see methods).
-
Figure 3—figure supplement 1—source data 1
Unedited agarose gel for Figure 1—figure supplement 1C.
- https://cdn.elifesciences.org/articles/87357/elife-87357-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Table related to Figure 3—figure supplement 1D.
- https://cdn.elifesciences.org/articles/87357/elife-87357-fig3-figsupp1-data2-v1.zip
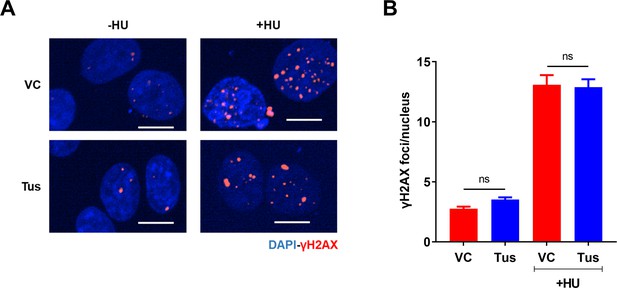
Genome-wide gH2AX foci were unaffected with Tus expression.
(A) Representative images of gH2AX foci across stated conditions. Scale Bars, 10 μm. (B) Average number of gH2AX foci per nucleus in conditions stated. Cells treated with or without 2 mM hydroxyurea (HU) for 4 hr (n = 3, ≥300 cells per experiment) ns: non-significant. Bar graphs represents mean ± standard error of the mean (SEM), Student t-test (see methods).
-
Figure 3—figure supplement 2—source data 1
Table related to Figure 3—figure supplement 2B.
- https://cdn.elifesciences.org/articles/87357/elife-87357-fig3-figsupp2-data1-v1.zip
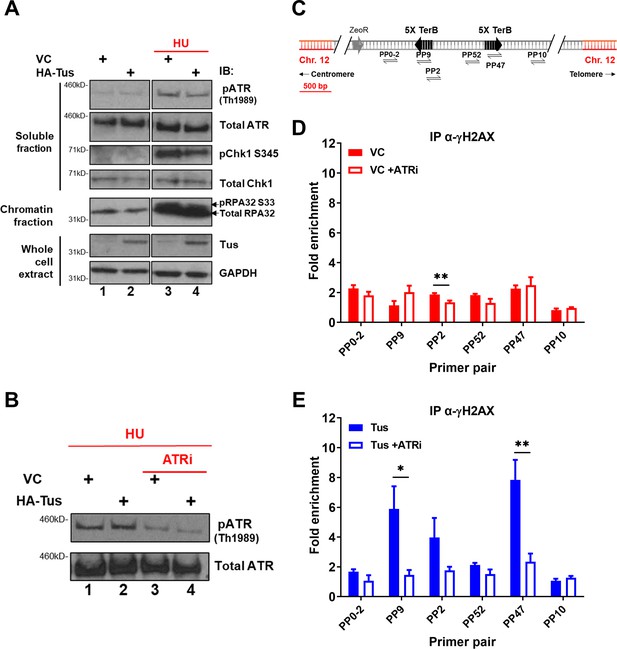
γH2AX phosphorylation at Tus-TerB is ATR-dependent.
(A) MCF7 5C-TerB cells transfected with vector control (VC) or HA-Tus were treated for 4 hr with 2 mM hydroxyurea (HU) before lysis and fractionation (whole-cell extract [WCE], soluble fraction, and chromatin fraction). Total and phosphorylated protein levels were examined by immunoblotting as indicated. Tus expression was confirmed in WCE. (B) Depletion of pATR Th1989 in MCF7 5C-TerB cells treated with 2 mM HU ± 40 nM of ATR inhibitor was examined with immunoblotting. (C) Schematic of MCF7 5C-TerB depicting the positions of the primer pairs with respect to the integrated TerB locus. (D, E) γH2AX levels were analyzed by ChIP-qPCR in MCF7 5C-TerB cells transfected with (D) VC expression plasmid (E) Tus expression plasmid, ±4 hr treatment with ATRi with indicated primer pairs. Data shows the fold enrichment over the IgG controls (n = 3). Bar graphs represents mean ± standard error of the mean (SEM), Student t-test (see methods).
-
Figure 4—source data 1
Unedited western blot images for Figure 4A.
- https://cdn.elifesciences.org/articles/87357/elife-87357-fig4-data1-v1.zip
-
Figure 4—source data 2
Unedited western blot images for Figure 4B.
- https://cdn.elifesciences.org/articles/87357/elife-87357-fig4-data2-v1.zip
-
Figure 4—source data 3
Tables related to Figure 4D and E.
- https://cdn.elifesciences.org/articles/87357/elife-87357-fig4-data3-v1.zip
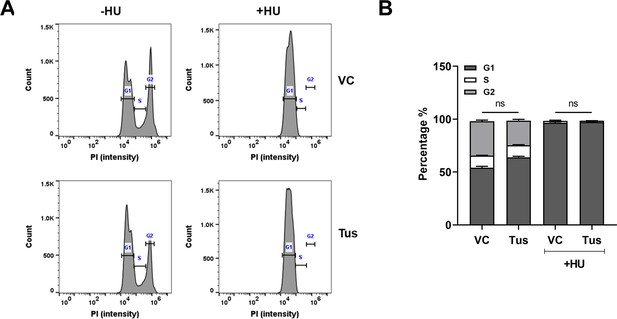
Cell cycle progression was unaffected by Tus expression.
(A) Cell cycle analysis and (B) quantification was performed using flow cytometry following staining with propidium iodide staining in MCF7 5C-TerB cells under the stated conditions. Cells treated with or without 2 mM hydroxyurea (HU) for 4 hr (n = 3, ns: non-significant). Bar graphs represents mean ± standard error of the mean (SEM), Student t-test (see methods).
-
Figure 4—figure supplement 1—source data 1
Table related to Figure 4—figure supplement 1B.
- https://cdn.elifesciences.org/articles/87357/elife-87357-fig4-figsupp1-data1-v1.zip
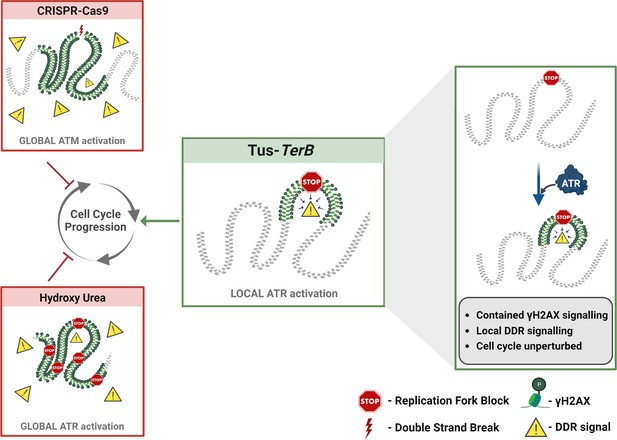
A local ATR-dependent checkpoint is activated by the Tus-TerB replication fork barrier (RFB).
A model depicting the cellular response to a site-specific DSB using CRISPR-Cas9 and a global replication stress with hydroxyurea, both of which lead to a DNA damage response (DDR, yellow triangles), increased gH2AX (green) levels globally in the cell and, if left unresolved, can alter cell cycle progression. In contrast, the site-specific replication fork block, Tus/TerB, elicits a local ATR-dependent DDR, which is responsible for the phosphorylation and accumulation of γH2AX at the stalled site. This local signaling does not affect the progression of the cell cycle and is not altered during the local checkpoint response.
Additional files
-
Supplementary file 1
Supplementary information for Figure 2 and extended methods.
(a) Demonstration that the Tus-Ter replication fork block does not activate significant replication elsewhere in the genome. Replication characteristics of 200 kb global DNA segments that represent the total genome. These measurements do not include the segments containing the TerB sequence. The table compares MCF7 cells containing the TerB sequence with and without Tus induced. This was determined on stretched DNA molecules that had completely incorporated IdU, CldU, or a combination of both nucleotide analogs. (b) List of antibodies, oligos, and plasmids used in the study.
- https://cdn.elifesciences.org/articles/87357/elife-87357-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/87357/elife-87357-mdarchecklist1-v1.pdf





