A positive feedback loop between ZEB2 and ACSL4 regulates lipid metabolism to promote breast cancer metastasis
Figures
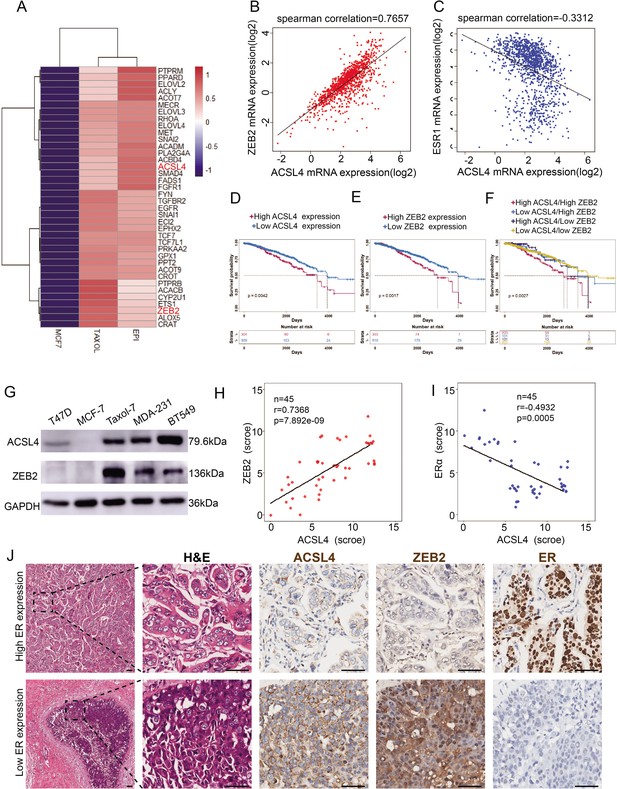
The expression and relationship of ZEB2 and ACSL4 in breast cancer.
(A) Heatmaps of the 38 upregulated epithelial–mesenchymal transition (EMT)-related genes detected by RNA-seq analysis in the paclitaxel-resistant MCF-7 cell line (TAXOL) and epirubicin-resistant MCF-7 cell line (EPI) compared to wild-type MCF-7 cell line. (B) The correlation between ACSL4 and ZEB2 mRNA expression in the TCGA cohort consisting of 1222 breast cancer patient samples. Spearman correlation and linear regression analysis were employed. (C) The correlation between ACSL4 and ERɑ mRNA expression in the TCGA cohort consisting of 1222 breast cancer patient samples. Spearman correlation and linear regression analysis were employed. (D) OS (overall survival) was examined by Kaplan–Meier analysis to compare the survival rates in ACSL high and low expression of breast cancer patients. (E) OS (overall-progression survival) as examined by Kaplan–Meier analysis to compare the survival rates in ZEB2 high and low expression of breast cancer patients. (F) OS (overall-progression survival) was examined by Kaplan–Meier analysis to compare the survival rates in the four groups of breast cancer patients. (G) Expression of ACSL4 and ZEB2 was analyzed by western blot in a panel of five breast cancer cell lines, including two basal-like (MAD-231, BT549), two luminal (T47D, MCF-7), and a Taxol-resistant cell lines. (H) The correlation between ACSL4 and ZEB2 protein expression in the immunohistochemistry (IHC) cohort consisting of 45 breast cancer patient samples. (I) The correlation between ACSL4 and ERɑ protein expression in the IHC cohort consisting of 45 breast cancer patient samples. (J) HE staining and IHC analysis of ACSL4, ZEB2, ERɑ expression in representative basal-like and luminal subtype breast cancer tissues. Representative pictures were shown. Scale bar, 50 μm.
-
Figure 1—source data 1
The gene expression datasets of the RNA-seq analysis of MCF-7 and EPI-resistant luminal breast cancer cell lines.
- https://cdn.elifesciences.org/articles/87510/elife-87510-fig1-data1-v1.xls
-
Figure 1—source data 2
The gene expression datasets of the RNA-seq analysis of MCF-7 and Taxol-resistant luminal breast cancer cell lines.
- https://cdn.elifesciences.org/articles/87510/elife-87510-fig1-data2-v1.xls
-
Figure 1—source data 3
The raw unedited gels or blots images of Figure 1.
- https://cdn.elifesciences.org/articles/87510/elife-87510-fig1-data3-v1.zip

The representative differential genes between wild-type MCF-7 cells, paclitaxel-resistant MCF-7 cells (TAXOL), and epirubicin-resistant MCF-7 cells (EPI).
(A) The heatmap of the upregulated genes in drug-resistant cells was shown. (B) The heatmap of the downregulated genes in the drug-resistant cell was shown.
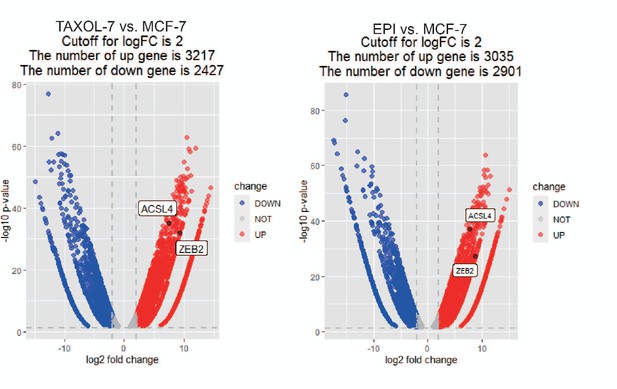
The volcano plot was generated using R4.3.0 software for differentially expressed genes in the paclitaxel-resistant MCF-7 cell line (TAXOL) and epirubicin-resistant MCF-7 cell line (EPI) compared to wild-type MCF-7 cell line.
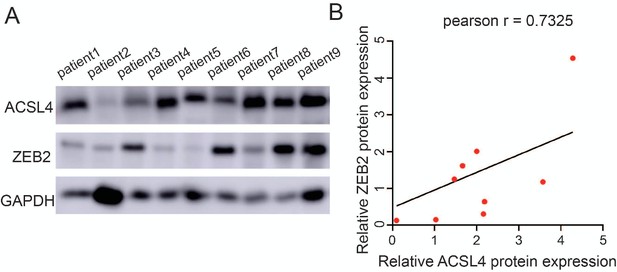
The protein expression of ACSL4 and ZEB2 in patients with breast cancer.
(A) Expression of ACSL4 and ZEB2 was analyzed by western blot in a panel of nine cases of tumor samples, including five cases of luminal (patients 1, 2, 3, 4, and 5) and four cases of triple-negative breast cancer (patients 6, 7, 8, and 9). (B) Scatter plot with linear regression analysis from expression level of A.
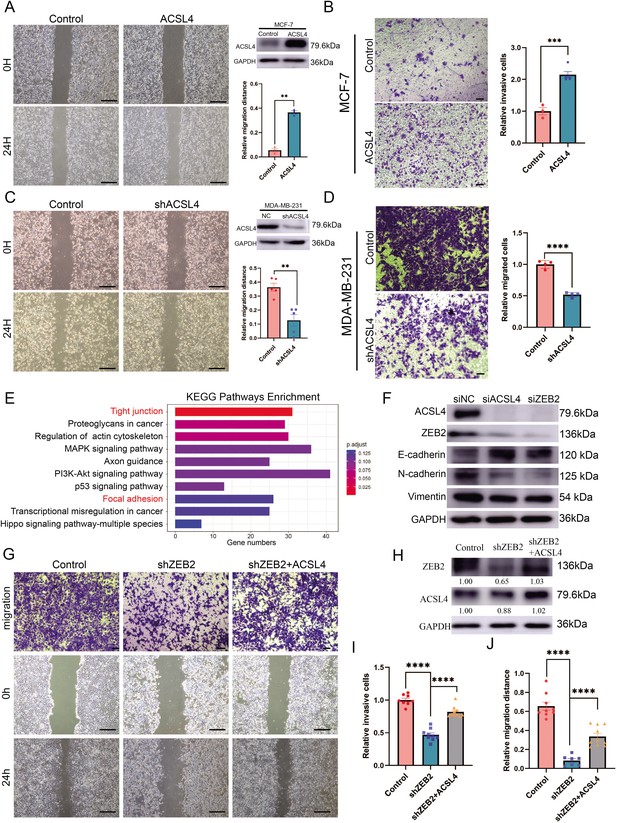
Overexpression of ACSL4 contributes to ZEB2-mediated breast cancer invasion.
(A) Cell metastatic capacity was analyzed by wound healing assay in control or ACSL4 overexpression MCF-7 cells (left panel). Expression of ACSL4 was analyzed by western blot in control or ACSL4 overexpression MCF-7 cells. Quantification of relative migration distance (right panel). Scale bar, 5 mm. (B) Cell invasive capacity was analyzed by transwell invasion assay in control or ACSL4 overexpression MCF-7 cells (left panel). Quantification of relative invasive cells (right panel). Scale bar, 1 mm. (C) Cell metastatic capacity was analyzed by wound healing assays in control or ACSL4 silencing MDA-MD-231 cells (shACSL4) (left panel). Expression of ACSL4 was analyzed by western blot in control or ACSL4 silencing MDA-MD-231 cells (shACSL4). Quantification of relative migration distance (right panel). Scale bar, 5 mm. (D) Cell invasive capacity was analyzed by transwell invasion assay in control or ACSL4 silencing MDA-MD-231 cells (shACSL4) (left panel). Quantification of relative invasive cells (right panel). Scale bar, 1 mm. (E) KEGG (https://www.kegg.jp/) pathway enrichment analysis of differentially expressed genes by RNA-sequencing between control and ACSL4 knockdown MDA-MB-231 cells. The top 10 deferential pathways were listed. (F) Expression of three epithelial–mesenchymal transition (EMT)-related genes, E-cadherin, N-cadherin, and vimentin, was analyzed by western blotting in control, ACSL4, or ZEB2-silencing MDA-MB-231 cells. (G) Cell invasive and metastatic capacities were analyzed by transwell invasion assay and wound healing assays (scale bar, 1/5 mm) in control and ZEB2 knockdown, or ZEB2 knockdown cells that overexpress ACSL4. (H) Protein expression was analyzed by western blot in control, ZEB2 knockdown (shZEB2), and ZEB2 knockdown with ACSL4 overexpression (shZEB2 + ACSL4) cells. (I) Quantification of relative invasive cells in G. (J) Quantification of relative migration distance in G. Graphs indicated the statistical analysis in G analyzed by Student’s t-test (mean ± standard error of the mean [SEM]). **p < 0.01, ***p < 0.001, ****p < 0.0001. All results are from three or four independent experiments.
-
Figure 2—source data 1
The datasets of differentially expressed genes by RNA-sequencing between control and ACSL4 knockdown MDA-MB-231 cells.
- https://cdn.elifesciences.org/articles/87510/elife-87510-fig2-data1-v1.xlsx
-
Figure 2—source data 2
The raw unedited gels or blots images of Figure 2.
- https://cdn.elifesciences.org/articles/87510/elife-87510-fig2-data2-v1.zip
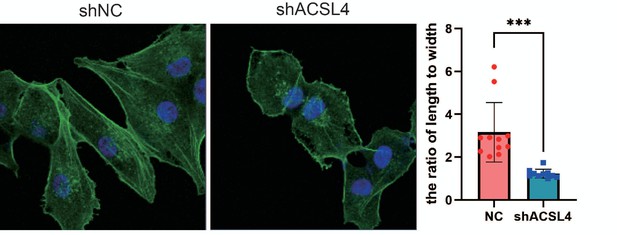
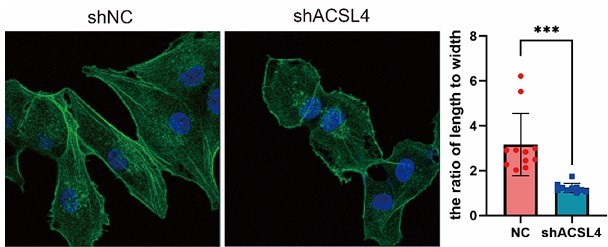
Fluorescence staining of Phalloidin in control or ACSL4 knockdown MDA-MB-231 cells.
The morphology of cells was analyzed by the ratio of length to width (right panel). ***p < 0.001.
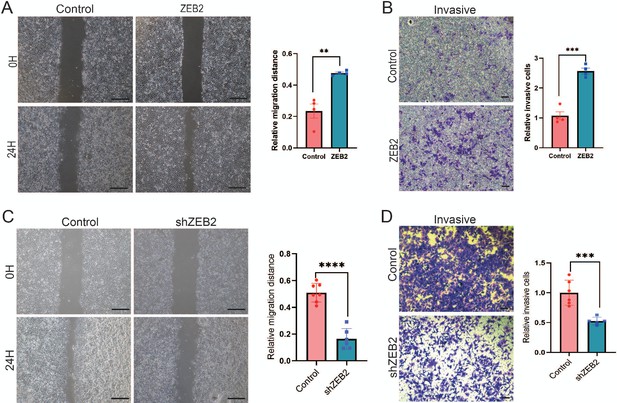
ZEB2 increases the metastatic and invasive capacities in breast cancer cells.
(A) Cell metastatic capacity was analyzed by wound healing assay in control or ZEB2 overexpression MCF-7 cells (left panel). Quantification of relative migration distance (right panel). Scale bar, 5 mm. (B) Cell invasive capacity was analyzed by transwell invasion assay in control or ZEB2 overexpression MCF-7 cells (left panel). Quantification of relative invasive cells (right panel). Scale bar, 1 mm. (C) Wound healing assays were performed in control cells or ZEB2 knockdown (shZEB2) MDA-MD-231 cells (left panel). Quantification of relative migration distance (right panel). Scale bar, 5 mm. (D) Transwell invasive assay was performed in control cells or ZEB2 knockdown (shZEB2) MDA-MD-231 cells (left panel). Quantification of relative invasive cells (right panel). Scale bar, 1 mm. **p < 0.01, ***p < 0.001, ****p < 0.0001.
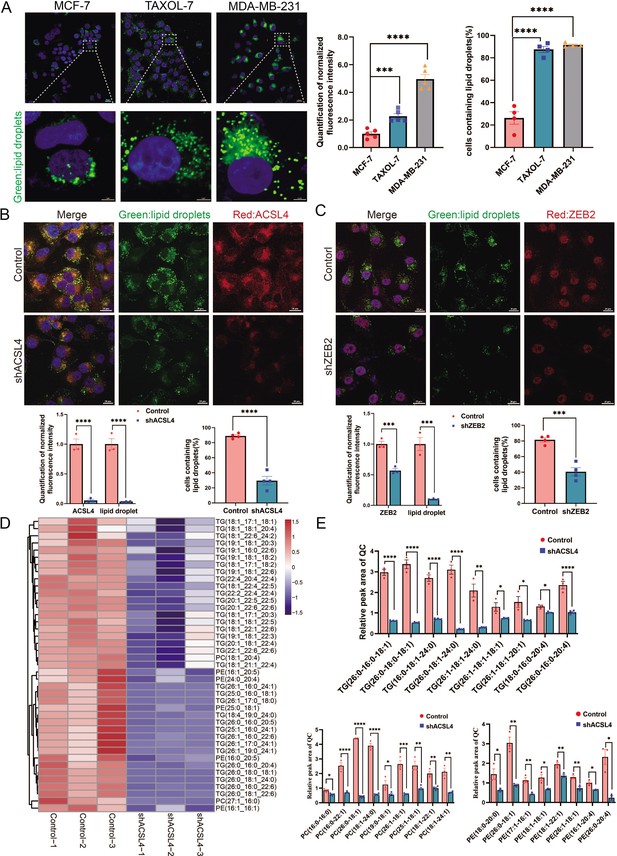
ACSL4 promotes lipid droplets (LDs) accumulation and lipogenesis.
(A) BODIPY 493/503 staining of LDs in MCF-7, Taxol-resistant MCF-7 cells (TAXOL-7), or MDA-MB-231 cells (left panel). Quantification of normalized lipid contents (right panel). Scale bar, 5 μm. (B) BODIPY 493/503 staining of LDs in control or ACSL4 knockdown MDA-MB-231 cells (upper panel). Quantification of normalized fluorescence intensity and percentage of LDs-containing cell number (lower panel). Scale bar, 50 μm. (C) BODIPY 493/503 staining of LDs in control or ZEB2 knockdown MDA-MB-231 cells (upper panel). Quantification of normalized fluorescence intensity and percentage of LDs-containing cell number (lower panel). Scale bar, 50 μm. (D) The heatmap of representative downregulated lipid species (TG, PE, and PC) with hierarchical clustering in the control cells and ACSL4 knockdown cells. Each species was normalized to the corresponding mean value, as determined by two-way analysis of variance (ANOVA). (E) Quantification of different fatty acids containing TG, PC, and PE species in control or ACSL4 knockdown MDA-MD-231 cells. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars, standard error of the mean (SEM).
-
Figure 3—source data 1
The lipidomics data between control and ACSL4 knockdown MDA-MB-231 cells.
- https://cdn.elifesciences.org/articles/87510/elife-87510-fig3-data1-v1.xlsx
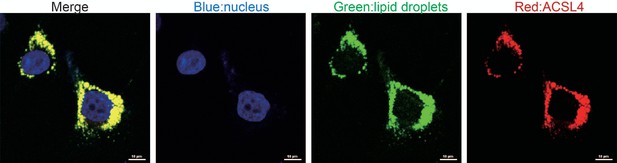
Fluorescence staining of lipid droplets and ACSL4 in MDA-MB-231 cells.
Scale bar, 10 µm.
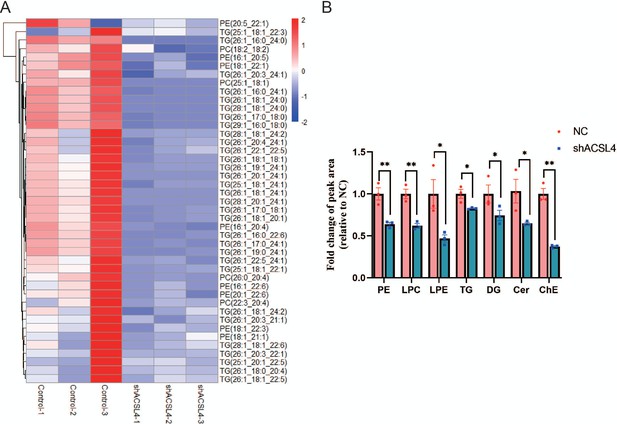
ACSL4 regulates lipid composition in basal-like breast cancer (BLBC) cells.
(A) The heatmap of the downregulated lipid species in ACSL4 knockdown cells (shACSL4) compared with those in control cells. (B) Quantification of different species of lipids in two groups as indicated. *p < 0.05, **p < 0.01.
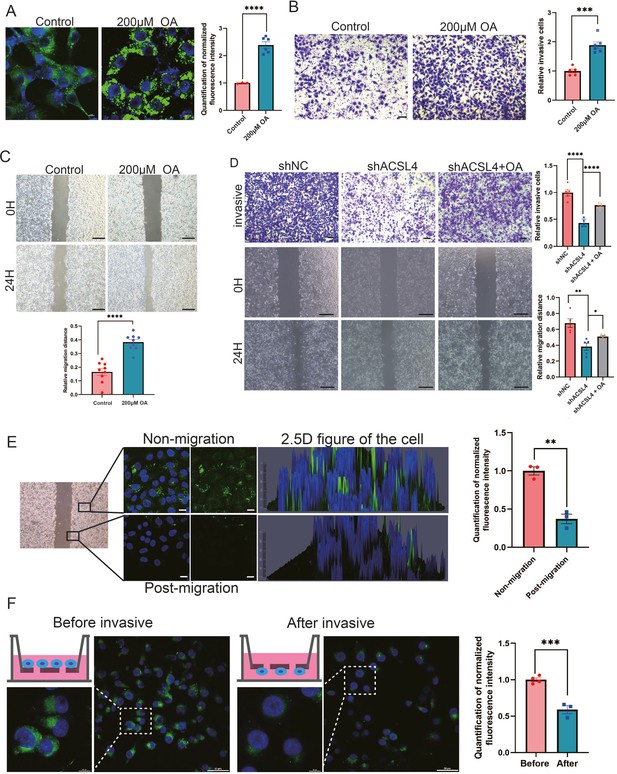
Exogenous lipids promote LDs accumulation and fuel cell migration.
(A) BODIPY 493/503 staining of LDs in control cells or oleic acid (OA) loaded (200 µM) MDA-MB-231 cells. Scale bar, 10 μm. Quantification of normalized lipid contents from the conditions in (A). (B) Cell invasive capacity was analyzed by transwell invasion assay in control or OA loaded (200 µM) MDA-MB-231 cells. Cells invaded for 16 hr through a Matrigel-coated filter toward high-serum media (left panel). Quantification of relative invasive cells (right panel). Scale bar, 1 mm. (C) Cell metastatic capacity was analyzed by wound healing assays in control or OA loaded (200 µM) MDA-MB-231 cells. Scale bar, 5 mm. (D) Cell invasive and metastatic capacities were analyzed by transwell invasion assay and wound healing assays in control, shACSL4, and OA loaded ACSL4 knockdown MDA-MB-231 cells. Quantification of relative invasive and migrated cells. Scale bar, 1 /5 mm. (E) BODIPY 493/503 staining of LDs in the cells at the leading edge of the scratch and the cells that away from the edge. The 2.5 D figure of the cell was shown. Quantitation of total LDs area per cell. Scale bar, 10 µm. (F) Cells were seeded in a transwell chamber. After 24 hr, cells migrated to the lower side of the chamber, and the fluorescence intensity per cell was calculated (left panel). Quantitation of total LDs area per cell before and after cell migration (right panel). Scale bar, 10 µm (small) and 50 µm (big). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
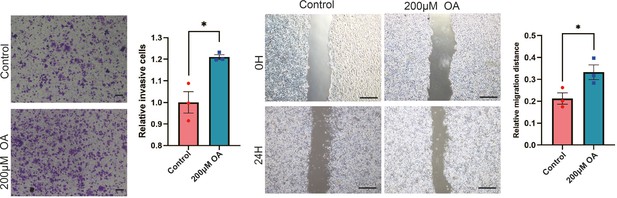
Oleic acid (OA) induces cell migration in breast cancer cells.
Cell invasive and metastatic capacities were analyzed by transwell invasion assay and wound healing assay in control or oleic acid (OA) loaded (200 μM) MCF-7 cells. Quantification of relative invasive cells (right panel). Scale bar, 1 mm (small), 5 mm (big). *p < 0.05.
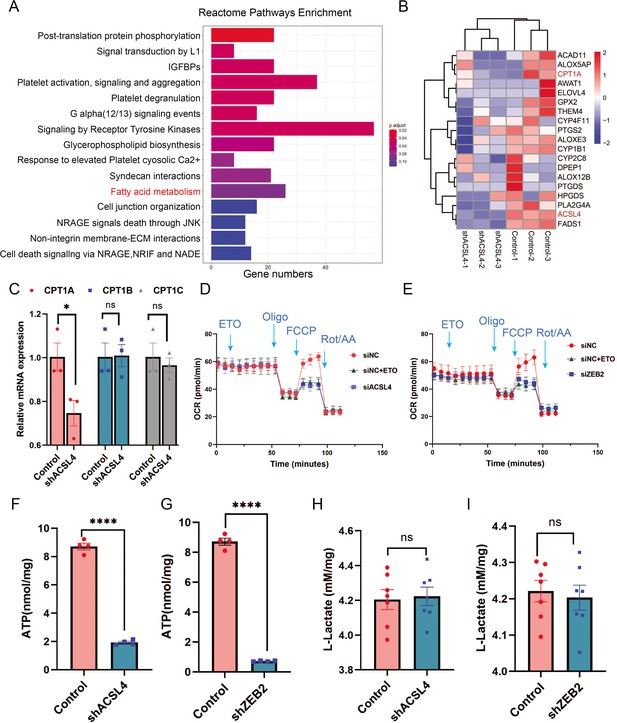
ACSL4 and ZEB2 increase fatty acid oxygen consumption and promote adenosine triphosphate (ATP) generation in basal-like breast cancer (BLBC) cells.
(A) Reactome pathway analysis of differentially expressed genes by RNA-sequencing between control and ACSL4 knockdown MDA-MB-231 cells. The representative pathways were shown. (B) Heatmap showing the differentially expressed genes between control cells and ACSL4 knockdown cells. The representative fatty acid oxidation (FAO)-related genes were shown. (C) The mRNA levels of CPT1A, CPT1B, and CPT1C were analyzed by quantitative PCR in control and ACSL4 knockdown cells. (D) Quantitation of the normalized oxygen consumption rate (OCR) for long-chain fatty acids was monitored by Agilent XF Substrate Oxidation Stress Test in control or ACSL4 knockdown cells. Specific inhibitors were added as indicated. (E) Quantitation of the normalized OCR for long-chain fatty acids was monitored by Agilent XF Substrate Oxidation Stress Test in control or ZEB2 knockdown cells. Specific inhibitors were added as indicated. (F, G) ATP production was quantified in control or ACSL4 knockdown cells (left) or ZEB2 knockdown cells (right). (H, I) Lactate production was examined in control and ACSL4 knockdown (shACSL4) or ZEB2 knockdown (shZEB2) cells. Data are represented as mean ± standard error of the mean (SEM) of three independent experiments, analyzed by Student’s t-test, *p < 0.05, ****p < 0.0001.
-
Figure 5—source data 1
The raw unedited gels or blots images of Figure 5.
- https://cdn.elifesciences.org/articles/87510/elife-87510-fig5-data1-v1.zip
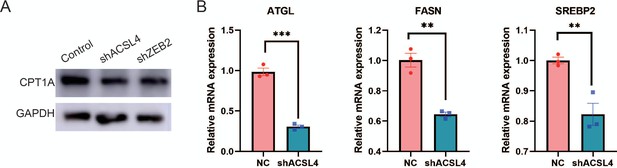
ACSL4 regulates the expression of lipid metabolic genes.
(A) Western blot analysis of CPT1A expression in control and ACSL4 knockdown, or ZEB2 knockdown MDA-MD-231 cells. (B) Relative mRNA levels of ATGL, FASN, and SREBP2 in control or ACSL4 knockdown MDA-MB-231 cells (shACSL4). **p < 0.01, ***p < 0.001.
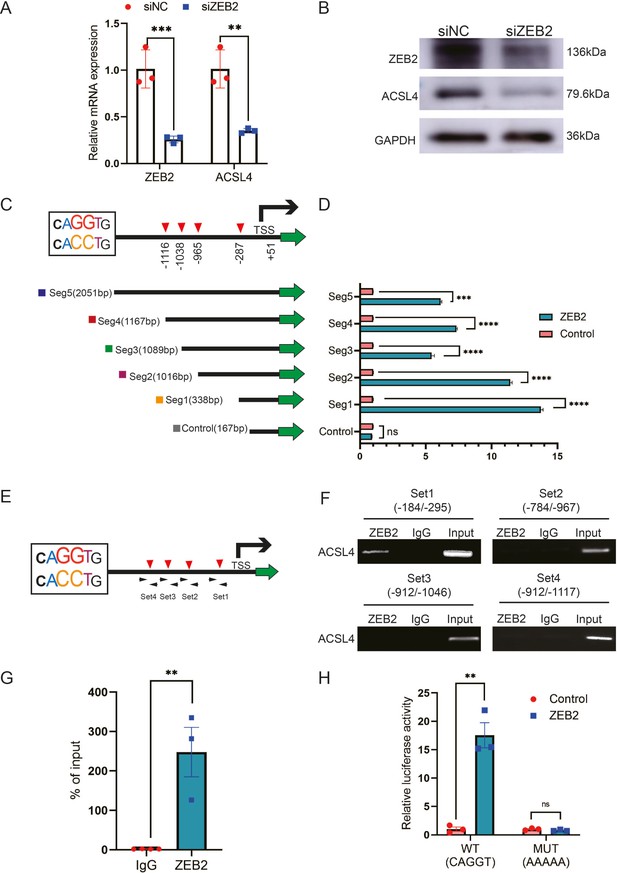
ZEB2 activates ACSL4 expression by directly binding to its promoter.
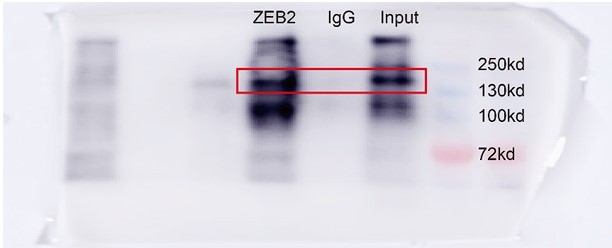
(A) Relative mRNA levels of ZEB2 and ACSL4 in control or ZEB2 knockdown MDA-MB-231 cells. (B) Protein levels of ZEB2 and ACSL4 in control or ZEB2 knockdown MDA-MB-231 cells. (C, D) Truncated ACSL4 promoter segment activity analyzed by luciferase reporter assay in control or ZEB2 overexpressed 293T cells. (E) The specific primers designed for ACSL4 promoter according to the E-box position shown in C (set1 to −287 bp, set2 to −965, set3 to −1038, set4 to −1116). (F) Chromatin immunoprecipitation (ChIP) assay analysis of the occupation of ZEB2 on ACSL4 promoter by using four primers as indicated in E. (G) Quantitative PCR analysis of ZEB2-binding abundance of specific ACSL4 promoter region by using set1 primer indicated in F. Genomic DNA was purified after ChIP and analyzed by quantitative PCR. (H) Luciferase reporter analysis of the activity of wild-type ACSL4 promoter or its mutants in control or ZEB2 overexpression 293T cells. Data are represented as mean ± standard error of the mean (SEM) of three independent experiments, analyzed by Student’s t-test, **p < 0.01, ***p <0 .001, ****p < 0.0001, ns: no significance.
-
Figure 6—source data 1
The raw unedited gels or blots images of Figure 6.
- https://cdn.elifesciences.org/articles/87510/elife-87510-fig6-data1-v1.zip
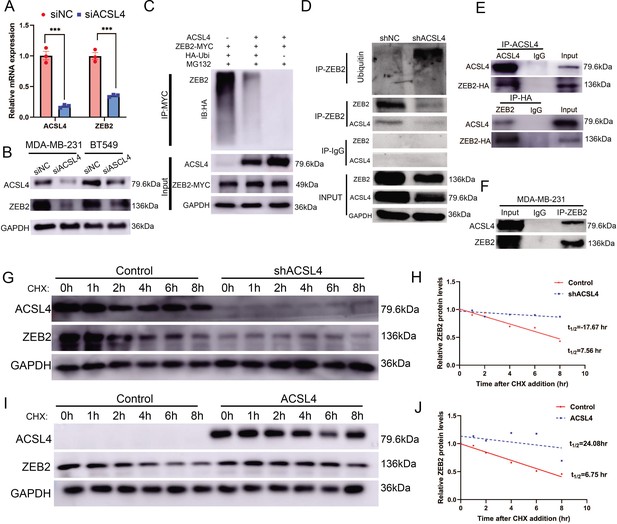
ACSL4 regulates ZEB2 mRNA expression and protein stability.
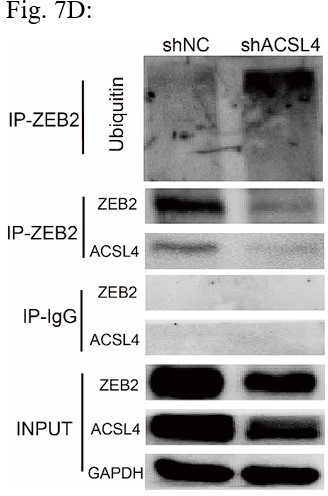
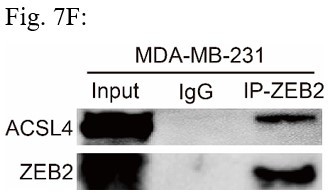
(A) Relative mRNA levels of ZEB2 and ACSL4 in control or ACSL4 silencing MDA-MB-231 cells. (B) Protein levels of ZEB2 and ACSL4 in control or ACSL4 silencing in MDA-MB-231 cells and BT549 cells as indicated. (C) Ubiquitylation of ZEB2 was examined by in vitro ubiquitin assay. 293T cells were co-transfected with indicated constructs. Cells were treated with MG132 for 6 hr before IP. Anti-MYC was used to pull down the ZEB2 protein. The polyubiquitinated ZEB2 protein was detected by an anti-HA antibody. (D) Ubiquitylation of ZEB2 was examined in MDA-MB-231 cells. The indicated antibody was used to pull down the protein in control or ACSL4 knockdown MDA-MB-231 cells. The polyubiquitinated ZEB2 protein was detected by anti-ubiquitin antibody. (E) The interaction between ACSL4 and ZEB2 was detected by co-immunoprecipitation (Co-IP) assay. 293T cells were co-transfected with ZEB2 and ACSL4 expressing construct. The indicated antibody was used to pull down the protein. (F) The interaction between ACSL4 and ZEB2 was detected by IP assay in MDA-MB-231 cells. Anti-ZEB2 antibody was used to pull down the protein. (G) The stability of ZEB2 protein was detected by CHX treatment assay in control or ACSL4 silencing MDA-MB-231 cells. Cells were treated with 100 µg/ml cycloheximide (CHX) and were harvested at the indicated times after the addition of CHX. GAPDH was used as the internal loading control. (H) Quantification of stability assays shown in G. (I) The stability of ZEB2 protein was detected by CHX treatment assay in control or ACSL4 overexpressed MCF-7 cells. GAPDH was used as the internal loading control. (J) Quantification of stability assays shown in I. ***p < 0.001.
-
Figure 7—source data 1
The raw unedited gels or blots images of Figure 7.
- https://cdn.elifesciences.org/articles/87510/elife-87510-fig7-data1-v1.zip
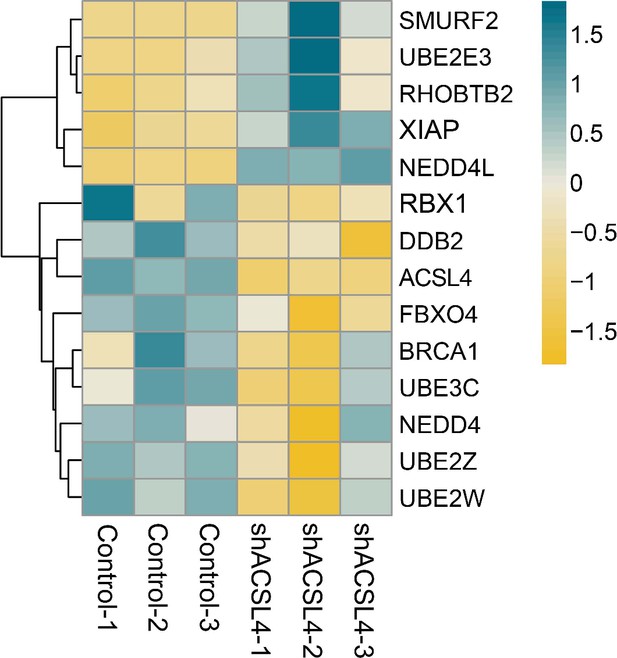
The differentially expressed genes between control cells and ACSL4 knockdown cells were shown.
The representative ubiquitination related proteolysis pathway genes were shown.
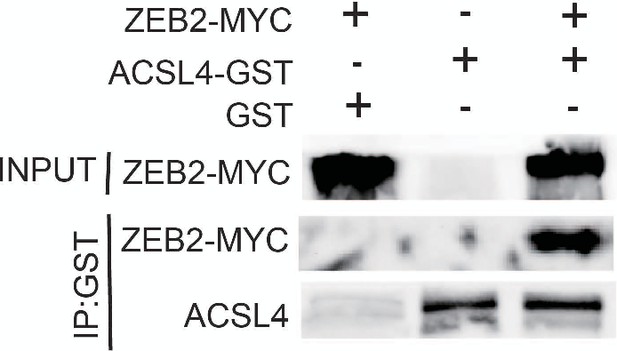
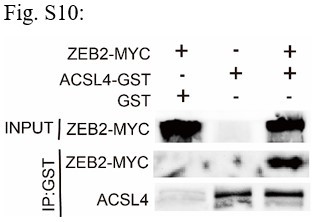
GST pull down assay was used to examine the interaction between ACSL4 and ZEB2 in MDA-MB-231 cells.
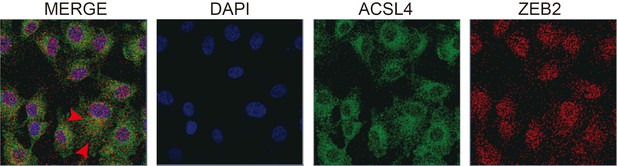
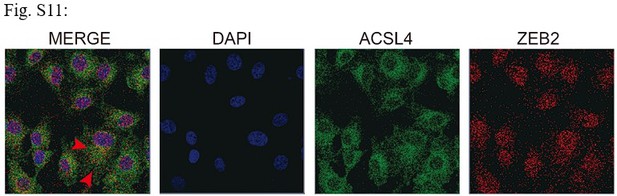
Fluorescence staining of ACSL4 and ZEB2 in MDA-MB-231 cells.
Scale bar, 10μM.
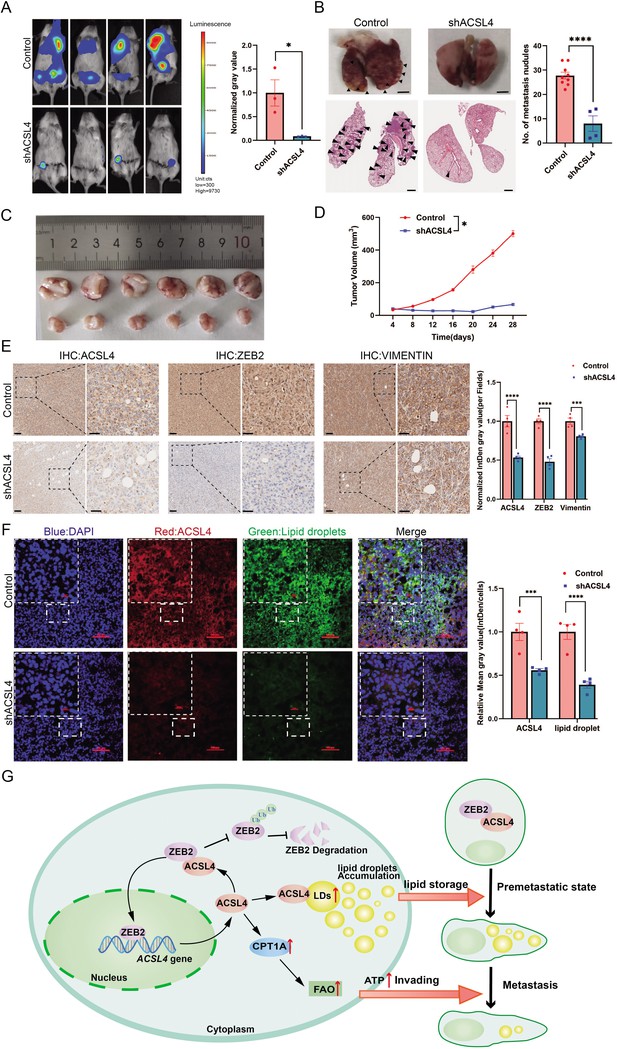
ACSL4 knockdown attenuates lung metastasis of breast cancer.
(A) Cell metastatic capacity was determined by xenograft experiment in vivo. Control cells and ACSL4 knockdown MDA-MB-231 cells were mixed with Matrigel in a 1:1 ratio and injected into NSG mice. Lung metastatic burden in mice was quantified by bioluminescence imaging at the experimental endpoint (left panel). Quantification of normalized gray area (right panel). (B) Representative xenograft tumor images and HE staining pictures of metastatic nodules in lungs (pointed by black arrows) were shown. The right panel shows the quantification of metastatic lung nodules in two groups as indicated. (C) The image of xenograft tumors developed from the control and ACSL4 knockdown cells. (D) The tumor volumes were measured and calculated at the indicated time. (E) Representative immunohistochemistry (IHC) images of lung metastatic lesions (left panel) and quantification is shown (right panel). Scale bar, 50 μm. (F) The representative images of fluorescence assay for ACSL4 and lipid droplets (left panel). The fluorescence intensity of lipid droplets and ACSL4 were calculated in the right panel. Scale bar, 100 μm. (G) A proposed mechanism to illustrate the positive feedback loop of ZEB2 and ACSL4 regulates lipid metabolism, which results in enhanced breast cancer metastasis. Data are represented as mean ± standard error of the mean (SEM) of three independent experiments and analyzed by Student’s t-test, *p < 0.05, ***p < 0.001, ****p < 0.0001.
Additional files
-
Supplementary file 1
The sequences of siRNA target.
- https://cdn.elifesciences.org/articles/87510/elife-87510-supp1-v1.docx
-
Supplementary file 2
The sequences of gene-specific primers used for quantitative reverse transcriptase polymerase chain reaction (qRT-PCR).
- https://cdn.elifesciences.org/articles/87510/elife-87510-supp2-v1.docx
-
Supplementary file 3
The control/−287 bp/−965 bp/−1036 bp/−1116 bp and –2000 bp regions and motif1 sequences of primers used for the ACSL4 promoter vector constructs.
- https://cdn.elifesciences.org/articles/87510/elife-87510-supp3-v1.docx
-
Supplementary file 4
The sequences of gene-specific primers used for chromatin immunoprecipitation (ChIP) assay.
- https://cdn.elifesciences.org/articles/87510/elife-87510-supp4-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/87510/elife-87510-mdarchecklist1-v1.docx