Genetic and dietary modulators of the inflammatory response in the gastrointestinal tract of the BXD mouse genetic reference population
Figures
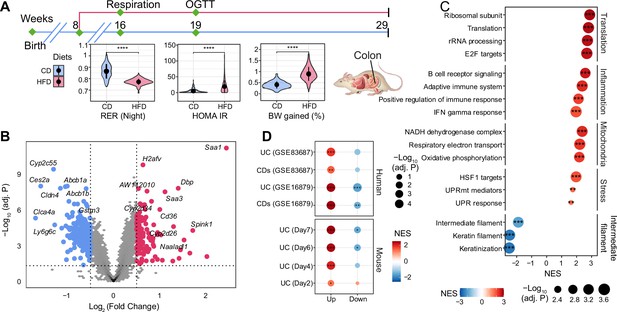
The effect of long-term high-fat diet (HFD) on gene expression in BXD colons.
(A) Graphical representation of a pipeline of a previously described BXD mouse study (Williams et al., 2016). Mice were fed HFD or chow diet (CD) starting from 8 wk of age, and metabolic phenotyping was performed as indicated. Mice were sacrificed at 29 wk of age, and multiple organs were collected and frozen for further analyses. BXD colon transcriptomes were analyzed in this study. CD is indicated in blue, and HFD is indicated in red. p-Values were calculated by two-tailed Student’s t-test and indicated as follows: *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. (B) Volcano plot showing the HFD effect on BXD colon transcriptomes compared to CD and the up- and downregulated differentially expressed genes (DEGs, absolute log2(fold change) > 0.5 and Benjamini–Hochberg [BH]-adjusted p-value (adj. p<0.05)) are highlighted in red and blue, respectively. (C) Gene set enrichment analysis showing the effect of HFD on gene expression in BXD colons. Gene sets were grouped into five categories: translation, inflammation, mitochondria, stress, and intermediate filament. Normalized enrichment scores (NES) are represented by color, and -Log10 (BH-adj. P) are represented by dot size and indicated as follows: *adjusted p-value<0.05; **adjusted p-value<0.01; ***adjusted p-value<0.001. (D) Enrichment analysis of molecular signatures of mouse and human inflammatory bowel disease (IBD) on the transcriptome of BXD colons. UC: ulcerative colitis; CDs: Crohn’s disease.
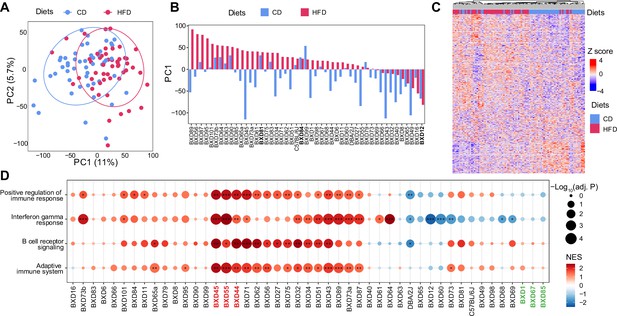
Transcriptome profile in BXD colons.
(A) Principal component analysis (PCA) of the microarray profiles of BXD colons under high-fat (HFD, indicated in red) or chow diet (CD, represented in blue). (B) Bar plot showing the primary principle (PC1) calculated by PCA of the colon transcriptomes in each BXD strain fed with CD (blue) or HFD (red). BXD strains highlighted in bold mean they are more resistant to dietary challenges. (C) Heatmap showing unsupervised hierarchical clustering of colon transcriptome in both the CD and HFD-fed BXDs. (D) Enrichment analysis of inflammation-related genesets showing the effect of HFD on gene expression in individual BXD colon. Normalized enrichment scores (NES) are represented by color, and -Log10(BH-adjusted p-values) are represented by dot size and indicated as follows: *adjusted p-value<0.05; **adjusted p-value<0.01; ***adjusted p-value<0.001. BXD strains highlighted in red represent the three most susceptible strains to gut inflammation upon HFD. BXD strains colored in green show no significant enrichment in gut inflammation.
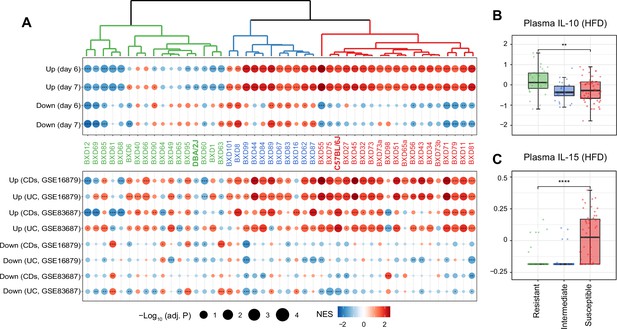
Identifying susceptible strains to high-fat diet (HFD)-induced inflammatory bowel disease (IBD)-like inflammation and their effect on plasma cytokines.
(A) Enrichment analysis of the molecular signatures in human (bottom panel) and mouse IBD (top panel) models on the gene expression of individual BXD colons. Dendrogram showing the propensity to develop a mouse IBD-like signature among BXD strains upon HFD. BXD strains were divided into three clusters: susceptible (in red), intermediate (in blue), and resistant strains (in green). Normalized enrichment scores (NES) are represented by color, and -Log10(BH-adjusted p-values) are represented by dot size and indicated as follows: *adjusted p-value<0.05; **adjusted p-value<0.01; ***adjusted p-value<0.001. UC: ulcerative colitis; CDs: Crohn’s disease. (B, C) Boxplots showing the effect of susceptible strains on plasma IL-10 (B) and IL-15 (C) level compared to resistant strains. p-Values were calculated by two-tailed Student’s t-test and indicated as follows: *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
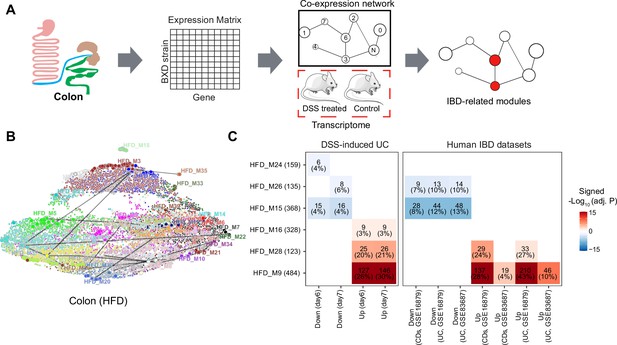
Identifying inflammatory bowel disease (IBD)-related gene modules.
(A) Pipeline for exploring the IBD-associated gene modules. A co-expression gene network was constructed based on the transcriptome of BXD colons under high-fat diet (HFD). IBD-associated modules were then defined as gene modules under HFD that are significantly clustered in mouse dextran sulfate sodium (DSS)-induced ulcerative colitis (UC) signatures. (B) Uniform Manifold Approximation and Projection (UMAP) representation of the co-expression gene network under HFD. Thirty-nine co-expression modules are represented in the corresponding color, and the correlated modules (Spearman correlation coefficient between the eigengene of modules >0.7) are linked by a gray line. (C) Heatmap showing the enrichment of co-expression modules in mouse and human IBD gene signatures, and the number and percentage of enriched genes are labeled. The number of enriched genes divided by the number of genes involved in the respective module is defined as the percentage of enriched genes. Signed –Log10(adj. p) indicates Benjamini–Hochberg (BH)-adjusted p-value and the enriched gene set. Enriched modules of up- and downregulated genes upon disease are highlighted in red and blue, respectively. CDs: Crohn’s disease.
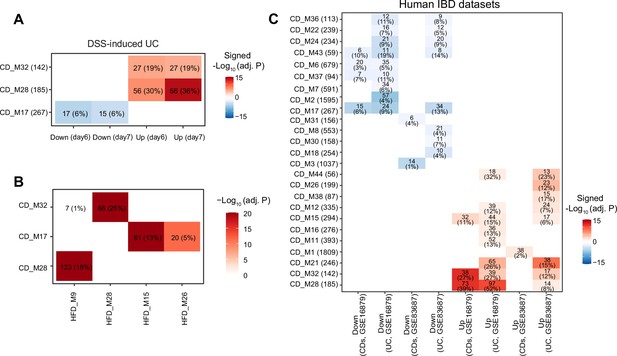
Exploring inflammatory bowel disease (IBD)-associated co-expression modules under chow diet (CD).
(A) Heatmap showing modules enriched in mouse IBD gene signatures, and the number and percentage of enriched genes are labeled. The number of enriched genes divided by the number of genes involved in the respective module is defined as the percentage of enriched genes. Signed –Log10(adj. p) indicates Benjamini–Hochberg (BH)-adjusted p-value and the enriched gene set. Enriched modules of up- and downregulated genes upon disease are highlighted in red and blue, respectively. (B) Heatmap showing the similarity of co-expression modules identified under CD or high-fat diet (HFD). The number and percentage of overlapped genes are labeled. The number of overlapped genes divided by the number of genes involved in the respective module in HFD is defined as the percentage of overlapped genes. Adjusted p-values calculated by BH are indicated by color. (C) Heatmap showing the modules under CD enriched in human IBD gene signatures, and the number and percentage of enriched genes are labeled. UC: ulcerative colitis; CDs: Crohn’s disease.

Biological interrogation of identified inflammatory bowel disease (IBD)-related modules.
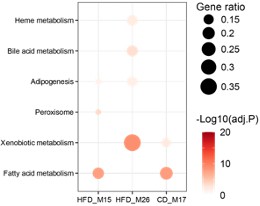
(A, B) Dotplots showing the enrichment of IBD-related modules in hallmark genesets (A) and cell-type gene signatures of inflamed colon in Crohn’s disease patients (B). Gene ratios higher than 0.1 are shown and represented by dot size. Dots are colored by -log10(BH-adjusted p-values). (C, D) The enriched motifs for promoters of the genes involved in module HFD_M9 (C) and HFD_M28 (D). The significantly enriched motifs (p-value<0.001) were ranked based on the percentage of enriched promoters (In top motifs) and then the top five TFs were selected. TF: transcription factor; PWM: positional weight matrix.
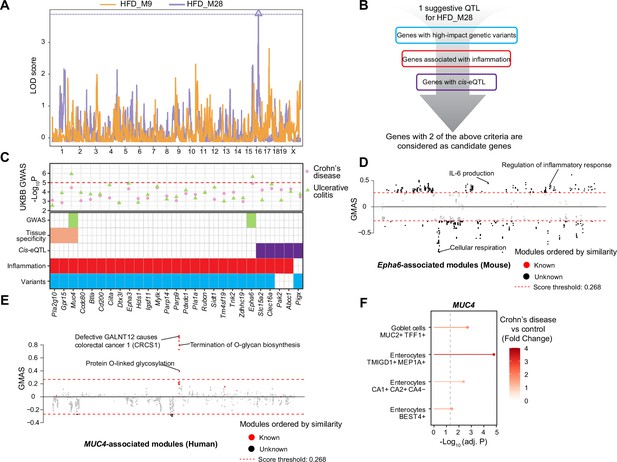
Module quantitative trait locus (ModQTL) mapping for two inflammatory bowel disease (IBD)-related modules and the prioritization of candidate genes.
(A) Manhattan plot showing the ModQTL mapping result for disease-related modules HFD_M9 and HFD_M28. ModQTL maps of HFD_M9 and HFD_M28 are indicated in orange and purple, respectively. The threshold calculated by permutation test (p<0.1) for HFD_M28 is represented by a purple dashed line. (B) The filtering criteria for selecting candidate genes under the ModQTL peak for HFD_M28. Genes with two of the described criteria are considered as candidate genes. (C) The most significant associations between 27 candidate genes under the ModQTL peak and Crohn’s disease or ulcerative colitis (UC) identified through genome-wide association studies (GWAS) according to whole-genome sequence in the human UK Biobank (UKBB) are shown in the scatter plot (top panel). Crohn’s disease and UC are indicated by pink circle and green triangle, respectively. The threshold (-Log10 p-value=5) is represented by a red dashed line. Heatmap showing the identified 27 candidate genes of module HFD_M28 (bottom panel). Variants colored in blue indicate genes with high-impact genetic variants in BXD mice (including missense, frameshift, initiator codon, splice donor, splice acceptor, in-frame deletion, in-frame insertion, stop lost, stop gained). Inflammation is indicated in red and represents genes associated with inflammation based on literature mining. Cis-eQTL colored in purple indicates genes with Cis-eQTLs. Tissue specificity colored in orange means genes that are highly expressed in human intestine (data were downloaded from human protein atlas, https://www.proteinatlas.org/humanproteome/tissue/intestine [Uhlén et al., 2015]). GWAS result of UC in UKBB colored in green indicates that genes are significantly associated with human UC. (D, E) Manhattan plots showing the associated gene expression modules of Epha6 in mouse gastrointestinal tract (D) and that of MUC4 in human gastrointestinal tract (E) (data from https://systems-genetics.org/gmad [Li et al., 2019]). The threshold is represented by the red dashed line (absolute Gene-Module Association Score [GMAS] ≥ 0.268). Terms above the threshold are identified as the significant associated terms. GO terms or gene modules are ranked by similarity. Known associated terms are shown as red dots, and new significant associated terms are colored in black. (F) Dot plot showing that the expression of MUC4 was higher in four cell types of human inflamed colon with Crohn’s disease.
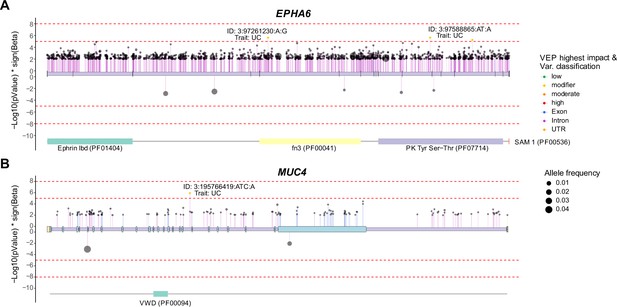
Genome-wide association studies (GWAS) for ulcerative colitis (UC) and Crohn’s disease (CDs) in humans.
(A, B) Lollipop plots showing UC- or CDs-associated genetic variants of EPHA6 (A) and MUC4 (B) based on UK Biobank (UKBB) whole-genome sequence data. Variants effect were predicted and their classification are represented by color. VEP: variant effect prediction.
Tables
Gene signatures of mouse and human IBDs.
| Disease | Source | DEGs | Thresholds | Species | Tissues |
|---|---|---|---|---|---|
| DSS-induced UC | GSE131032; Czarnewski et al., 2019 | Author-provided DEGs | Absolute log2 FC > 1 and BH-adjusted p-value<0.05 | Mouse | Colon |
| Crohn’s disease and UC | GSE16879; Arijs et al., 2009; Li et al., 2019 | Author-provided DEGs | Absolute FC > 1 and BH-adjusted p-value<0.01 | Human | Colon |
| Crohn’s disease and UC | GSE83687; Peters et al., 2017 | DEGs computed by limma package; Ritchie et al., 2015 | Absolute log2 FC > 1 and BH-adjusted p-value<0.001 | Human | Colon |
| Crohn’s disease | SCP1884; Kong et al., 2023 | Author-provided DEGs | DE coefficient > 1 and BH-adjusted p-value<0.05 | Human | Colon |
-
BH, Benjamini–Hochberg; DEGs, differentially expressed genes; DSS, dextran sulfate sodium; FC, fold change; IBD, inflammatory bowel diseases; UC, ulcerative colitis.
Additional files
-
Supplementary file 1
Co-expression networks under chow diet (CD) or high-fat diet (HFD).
Related to Figure 3.
- https://cdn.elifesciences.org/articles/87569/elife-87569-supp1-v1.xlsx
-
Supplementary file 2
Genes under quantitative trait locus (QTL) peak of module HFD_M28.
Related to Figure 5.
- https://cdn.elifesciences.org/articles/87569/elife-87569-supp2-v1.xlsx
-
Supplementary file 3
Associations between genetic variants of EPHA6 and inflammatory bowel disease (IBD).
Related to Figure 5 and its Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/87569/elife-87569-supp3-v1.xlsx
-
Supplementary file 4
Associations between genetic variants of MUC4 and inflammatory bowel disease (IBD).
Related to Figure 5 and its Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/87569/elife-87569-supp4-v1.xlsx
-
Supplementary file 5
G-MAD result for Epha6 in mice and MUC4 in humans.
Related to Figure 5
- https://cdn.elifesciences.org/articles/87569/elife-87569-supp5-v1.xlsx
-
Supplementary file 6
Mendelian randomization (MR) result for MUC4.
- https://cdn.elifesciences.org/articles/87569/elife-87569-supp6-v1.xlsx
-
Supplementary file 7
Medicine for human inflammatory bowel disease (IBD).
- https://cdn.elifesciences.org/articles/87569/elife-87569-supp7-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/87569/elife-87569-mdarchecklist1-v1.docx