A unique cell division protein critical for the assembly of the bacterial divisome
Figures
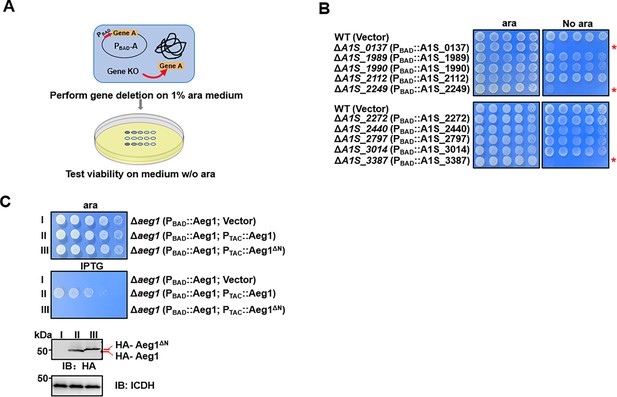
Identification of aeg1 as an essential gene for A. baumannii viability on rich medium.
(A) A diagram depicting the method for conditional deletion of potential essential genes. The gene to be examined was expressed from the arabinose-inducible promoter on a plasmid and the gene was deleted from the chromosome using the standard allele exchange method (upper panel) and strains lacking the chromosomal gene were tested for essentiality by spotting diluted cells on media with and without arabinose, respectively (lower panel). (B) Identification of A1S_0137, A1S_2249, and A1S_3387 as A. baumannii essential genes. Each of the 10 genes predicted to be essential were deleted by the method described in A and the resulting bacterial strains were tested for growth by spotting serially diluted cells on medium with or without 1% arabinose. Images were acquired after incubation at 37°C for 18 hr. Similar results were obtained in at least three independent experiments. (C) Cells harboring differentially regulated HA-Aeg1 and HA-Aeg1∆N spotted onto Luria Bertani (LB) agar supplemented with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) or 1% ara, images were acquired after 18 hr incubation at 37°C (C, upper two panels). The expression of the Aeg1 and Aeg1∆N were examined by immunoblotting with the HA-specific antibody. The metabolic enzyme isocitrate dehydrogenase (ICDH) was probed as a loading control (C, lower two panels). Similar results were obtained in three independent experiments. The red asterisk denotes that the deletion of this gene results in pronounced growth defects in the cells.
-
Figure 1—source data 1
PDF file containing original western blots for Figure 1C, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/87922/elife-87922-fig1-data1-v1.zip
-
Figure 1—source data 2
Original files for western blot analysis displayed in Figure 1C.
- https://cdn.elifesciences.org/articles/87922/elife-87922-fig1-data2-v1.zip
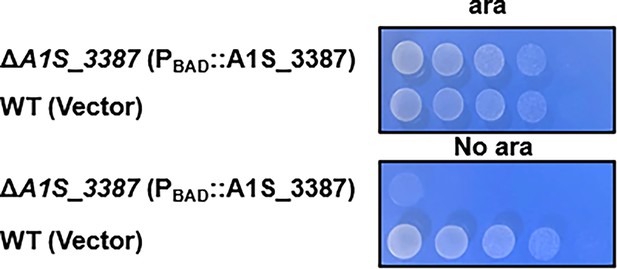
aeg1 is essential for A. baumannii growth in the VBS medium.
Strains were tested for growth by spotting serially diluted cells on medium with or without 1% arabinose. Images were acquired after incubation at 37°C for 18 hr. Similar results were obtained in at least three independent experiments.
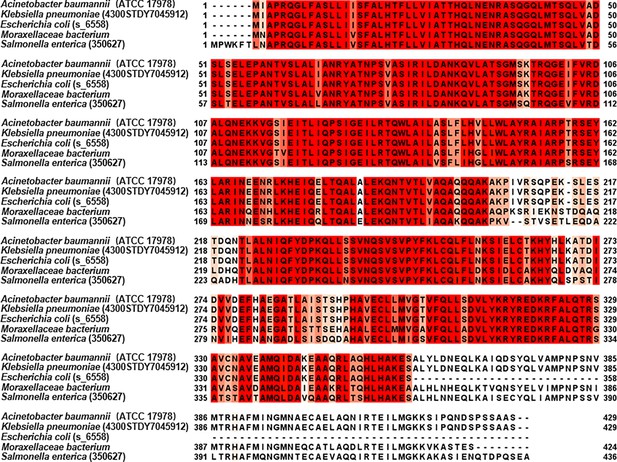
Alignment of Aeg1 homologs from a few Gram-negative bacteria.
Aeg1 of A. baumannii and its homologs from a member of the Moraxellaceae family, E. coli (strain s_6558), Klebsiella pneumoniae (strain 4300STDY7045912), and Salmonella enterica (strain 350627) were compared with Jalview. Homology from high to low was presented in dark red, orange, yellow, light yellow, or no highlight.
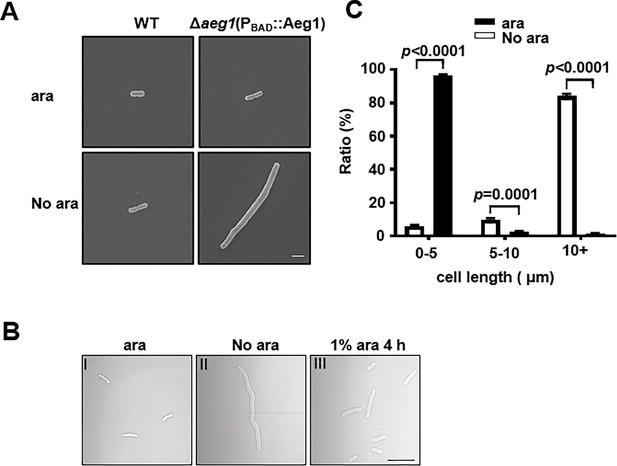
Aeg1 deficiency causes cell elongation in A. baumannii.
(A) Cell morphology of the wild-type and strain Δaeg1(PBAD::Aeg1) grown in medium with or without ara. The Δaeg1(PBAD::Aeg1) and the wild-type strains were grown in medium with or without ara for 18 hr, samples were processed for scanning electron microscope imaging. Images shown are representative of three parallel cultures. Bar, 2 µm. (B, C) Expression of Aeg1 reversed the cell elongation phenotype caused by its depletion. Saturated bacterial cultures of Δaeg1(PBAD::Aeg1) diluted in fresh Luria Bertani (LB) broth with (I) or without (II) ara were incubated for 18 hr. The culture from the uninduced sample was split into two subcultures, ara was added to one of the subcultures and the morphology of the cells was determined at 4 hr (III) post induction. Images are representative of three parallel cultures. Bar, 10 µm (B). Quantitation of the cell length in samples differently expressing Aeg1. Cell length was categorized into groups of 0–5 μm, 5–10 μm, and more than 10 μm. At least 300 cells were counted for each sample (C). Results shown were from the average of three independent experiments. Statistical analysis in each panel was performed by Student’s t-test.
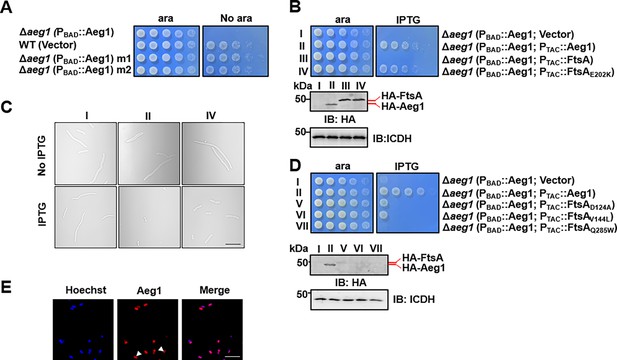
Mutations in proteins involved in cell division bypass the requirement of Aeg1.
(A) Isolation of two suppressor mutants that gained viability in the absence of Aeg1. Cells serially diluted with water were spotted onto Luria Bertani (LB) agar with or without ara. Note that the Δaeg1(PBAD::Aeg1) strain (top row) cannot grow on medium without ara and the wild-type strain (second row) can grow on both conditions. Similar results were obtained in three independent experiments. (B) The FtsAE202K mutant suppressed the requirement of Aeg1. Cells of strains derived from Δaeg1(PBAD::aeg1) that harbored the vector (I), pHA-Aeg1 (II), pHA-FtsA (III), or pHA-FtsAE202K (IV) were spotted onto LB agar containing ara (left) or isopropyl-β-D-thiogalactopyranoside (IPTG; right), Images were acquired after 18 hr incubation at 37°C (B, upper panels). Expression of Aeg1, FtsA, or FtsAE202K in these strains were detected by immunoblotting with the HA-specific antibody after lysates of IPTG-induced cells being resolved by SDS–PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis). Isocitrate dehydrogenase (ICDH) was probed as a loading control (B, lower panels). Similar results were obtained in three independent experiments. (C) The suppression mutants assumed normal cell morphology. Bacterial strains derived from Δaeg1(PBAD::aeg1) grown in LB broth containing ara for 6 hr were diluted into fresh medium with the inducer and the cultures were induced with IPTG for 4 hr prior to being processed for imaging. Images were representatives of three parallel cultures. Bar, 10 µm. (D) Additional FtsA mutants bypassed the need of Aeg1. Mutant FtsAD124A, FtsAV144A, or FtsAQ285A expressed from the IPTG-inducible PTAC was introduced into strain Δaeg1(PBAD::Aeg1) and serially diluted cells of resulting strains were spotted onto LB agar supplemented with ara or IPTG. Images were acquired after incubation at 37°C for 18 hr (upper panels). Expression of Aeg1, FtsAD124A, FtsAV144A, and FtsAQ285A in these strains. Total protein of IPTG-induced cells resolved by SDS–PAGE and proteins transferred onto nitrocellulose membranes were detected by immunoblotting with the HA-specific antibody. ICDH was probed as a loading control (lower panels). Similar results were obtained in three independent experiments. (E) The Aeg1-mCherry fusion localizes to division constrictions. Wild-type A. baumannii harboring PBAD::Aeg1-mCherry grown to the mid-log phase in LB broth containing 1% ara were processed for imaging. Bar, 10 µm. Images are representative of three parallel cultures.
-
Figure 3—source data 1
PDF file containing original western blots for Figure 3B, D, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/87922/elife-87922-fig3-data1-v1.zip
-
Figure 3—source data 2
Original files for western blot analysis displayed in Figure 3B, D.
- https://cdn.elifesciences.org/articles/87922/elife-87922-fig3-data2-v1.zip
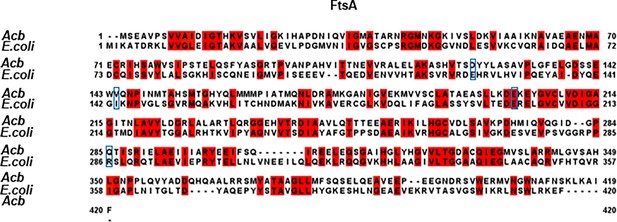
Alignment of FtsA proteins between A. baumannii and E. coli.
The FtsA proteins from A. baumannii and E. coli were compared using Jalview. The residues highlighted by blue boxes indicate the site of dominant active sites in E. coli proteins, and corresponding mutations were made in their A. baumannii counterparts.
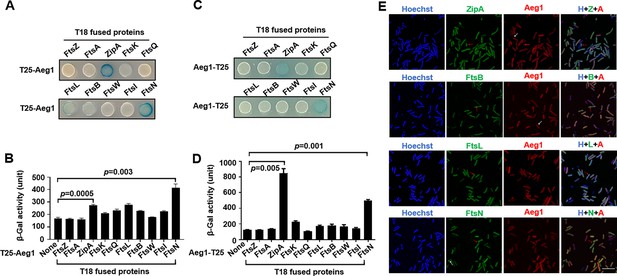
Aeg1 interacts with multiple core proteins of the A. baumannii divisome.
(A–D) Derivatives of strain E. coli BTH101 containing T18-Aeg1 and T25 fusion of the indicated proteins were spotted onto Luria Bertani (LB) agar containing X-gal. T25 domain was fused to the N-terminus (A, B) or C-terminus (C, D) of Aeg1. Images were acquired after incubation for 18 hr at 37°C (A, C). Interactions determined by quantitative measurement of β-galactosidase activity (B, D). Results shown were from three independent experiments. Statistical analysis was performed by Student’s t-test. (E) Aeg1 colocalized with a set of core divisome proteins. GFP(Green Fluorescent Protein). fusion of ZipA, FtsL, FtsB, or FtsN was expressed in strain WT(pJL03::mCherry-Aeg1). Saturated cultures of bacterial strains harboring the GFP and mCherry fusions were diluted into fresh LB broth. After the cell density of the subcultures reached until OD600 = 0.6, isopropyl-β-D-thiogalactopyranoside (IPTG) (0.25 mM) and ara (0.25%) were added to induce the expression of fusion proteins for 4 hr prior to being processed for imaging. Red arrows indicate the sites of colocalization. Cells in which the proteins did not colocalize were shown by white arrows. Bar, 10 µm. Images are representative of three independent cultures.
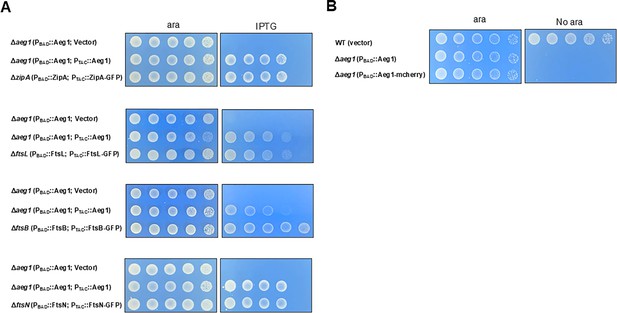
GFP and mCherry fusion proteins rescued the growth defects of their corresponding mutants.
zipA, ftsB, ftsL, and ftsN are essential cell division genes which were deleted by the method described in Figure 1A. The resulting bacterial strains were tested for growth by spotting serially diluted cells on medium with 1% arabinose or 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) (A). Strain Δaeg1(PBAD::mCherry-Aeg1) expressing mCherry fusion of Aeg1 from the PBAD promoter grown in medium with or without ara were processed for imaging (B). Images were acquired after incubation at 37°C for 18 hr. Similar results were obtained in at least three independent experiments.
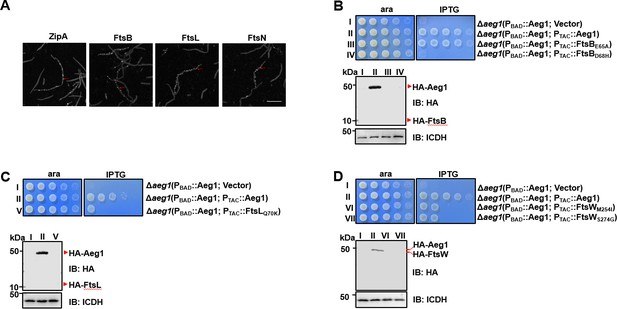
Aeg1 dictates the cellular localization of core divisome proteins.
(A) Aeg1 deficiency prevented core divisome proteins from localizing to the middle cell. Cells of derivatives of strain Δaeg1(pJL03::mCherry-Aeg1) expressing GFP fusion of each of the divisome proteins from the PTAC promoter grown in medium with ara were processed for imaging. The red arrows representative examples of the inability of fusion proteins to target to the septal ring. Bar, 5 µm. Images are representative of three parallel cultures. A few dominant active mutants of FtsB and FtsW bypassed the need of Aeg1 by A. baumannii. Mutant FtsBE65A (B), FtsBD68H (B), FtsLQ70K (C), FtsWM254I (D), or FtsWS274G (D) was expressed from the isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible promoter PTAC in strain Δaeg1(pJL03::Aeg1). Serially diluted cells were spotted onto Luria Bertani (LB) agar containing ara or IPTG. Images were acquired after incubation at 37°C for 18 hr (B–D). Similar results were obtained in three independent experiments. The expression of the protein was probed with the HA-specific antibody after IPTG induction (B–D). Isocitrate dehydrogenase (ICDH) was probed as a loading control.
-
Figure 5—source data 1
PDF file containing original western blots for Figure 5B–D, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/87922/elife-87922-fig5-data1-v1.zip
-
Figure 5—source data 2
Original files for western blot analysis displayed in Figure 5B–D.
- https://cdn.elifesciences.org/articles/87922/elife-87922-fig5-data2-v1.zip
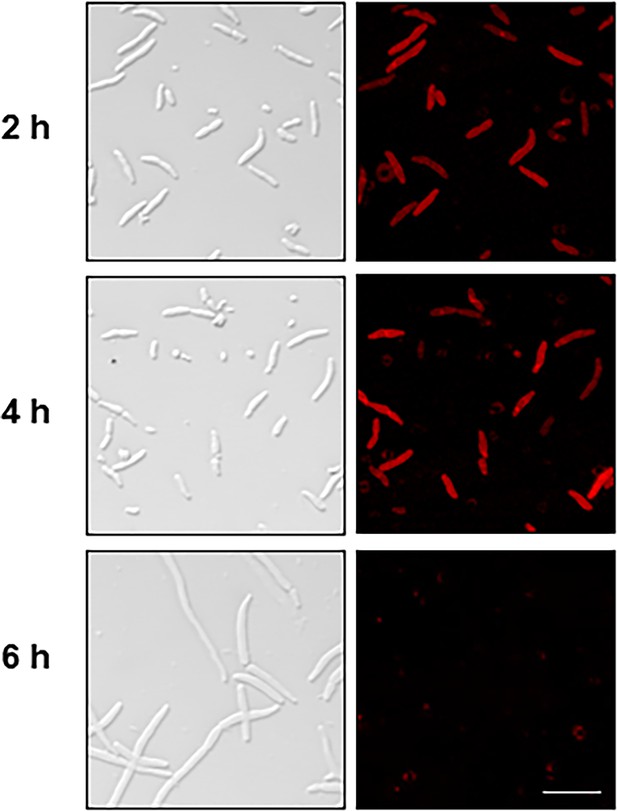
Δaeg1(PBAD::mCherry) strain unable to exhibit expression of Aeg1-mCherry after 6 hr period without ara.
Cells of derivatives of strain Δaeg1(PBAD::mCherry-Aeg1) expressing mCherry fusion of Aeg1 from the PBAD promoter grown in medium with ara were processed for imaging. Bar, 10 µm. Images are representative of three parallel cultures.
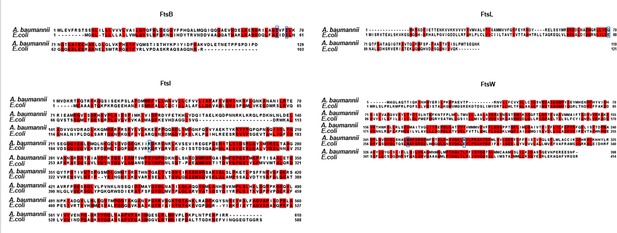
Alignment of Fts proteins between A. baumannii and E. coli.
The FtsB, FtsL, FtsW, and FtsI proteins from A. baumannii and E. coli were compared using Jalview. The residues highlighted by blue boxes indicate the site of dominant active sites in E. coli proteins, and corresponding mutations were made in their A. baumannii counterparts.
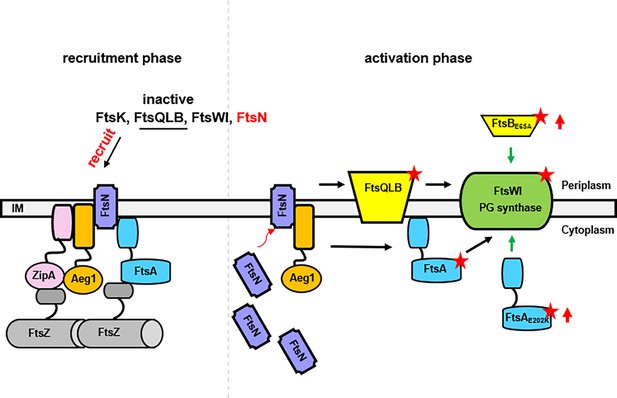
A model for the function of Aeg1 in cell division of A. baumannii.
In the early stages of Z-ring assembly, anchor proteins FtsA and ZipA tether FtsZ to the inner face of the plasma membrane. In the initial phase of cell division, a small amount of FtsN is recruited to the divisome in a FtsA-dependent manner (left, recruitment phase) (Wroblewska et al., 2007). The Aeg1–ZipA–FtsN complex acts as a dynamic scaffold for recruiting downstream of Fts proteins. Once more FtsN is recruited to the divisome, it will activate both FtsA and the FtsQLB complex, which in turn activates FtsI in the periplasm (Li et al., 2021). Note that constitutively active form of FtsA (FtsA*) acts on FtsW in the cytoplasm (Karimova et al., 1998) so did the hyperactive mutants FtsAE202K. These mutants all bypassed the need of Aeg1 (right, activation phase). Constitutively active mutants were indicated by a star. The red arrows next to FtsAE202K or FtsBE65A indicate overexpression of these mutants.
Additional files
-
Supplementary file 1
The essential genes identified and analyzed within this study.
- https://cdn.elifesciences.org/articles/87922/elife-87922-supp1-v1.docx
-
Supplementary file 2
A comparative analysis of substitution and insertion–deletion (InDel) mutations found in strains M1 and M2 relative to the genome sequence of A. baumannii strain ATCC 17978.
- https://cdn.elifesciences.org/articles/87922/elife-87922-supp2-v1.xls
-
Supplementary file 3
A comprehensive list of the bacterial strains, plasmids, and primers utilized throughout the study.
- https://cdn.elifesciences.org/articles/87922/elife-87922-supp3-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/87922/elife-87922-mdarchecklist1-v1.pdf





