Mouse gingival single-cell transcriptomic atlas identified a novel fibroblast subpopulation activated to guide oral barrier immunity in periodontitis
Figures
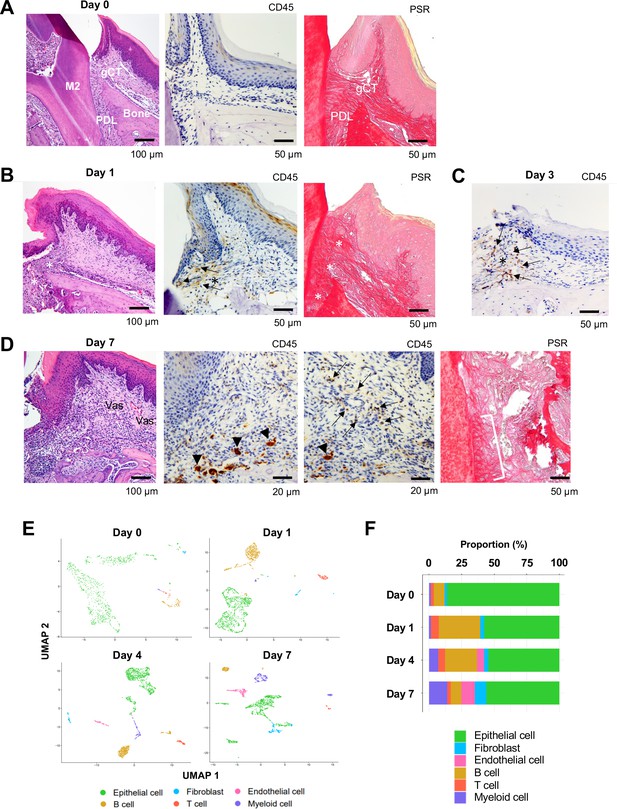
Changes in proportions of major cell types during ligature-induced periodontitis development in mice.
(A) Mouse maxillary second molar (M2) has multi-roots and supported by alveolar bone (Bone), periodontal ligament (PDL), and gingiva connective tissue (gCT). The oral barrier immunity is not constitutively activated as evidenced by the lack of CD45+ immune cells. Picrosirius red (PSR)-stained collagen fibers connected the root surface and alveolar bone in the PDL and organized as dense parallel bundles in gCT. (B) One day (day 1) after a ligature (5.0 silk suture) was placed around M2, the cervical PDL and gCT demonstrated a localized connective tissue degradation (*), where CD45+ immune cells clustered (arrows). PSR-stained collagen architecture immediately under the ligature lost the thick collagen bundle structure (*). (C) Day 3 of ligature placement exhibited localized but increased CD45+ immune cell clustering adjacent to the collagen degradation area (*). (D) Day 7 of ligature placement, PDL, and gCT tissue degradation increased with inflammatory vascularization (Vas). CD45+ myeloid cells (arrowheads) were observed near the alveolar bone surface and CD45+ lymphocytes (arrows) infiltrated the gCT area. PSR staining lost the typical collagen pattern in gCT and PDL, and a remnant of degraded PDL collagen fiber (white bracket) was attached to the tooth surface. (E) Single-cell RNA sequencing (scRNA-seq) t-distributed stochastic neighbor embedding (t-SNE) projection plots showing the major cell types present within gingival tissue during periodontitis development on days 0, 1, 4, and 7. Colors indicate cell type as follows: green, epithelial cells; blue, fibroblasts; pink, endothelial cells; yellow, B cells; red, T cells; and purple, myeloid cells. (F) Proportion plots showing the relative amounts of each major cell type on days 0, 1, 4, and 7.
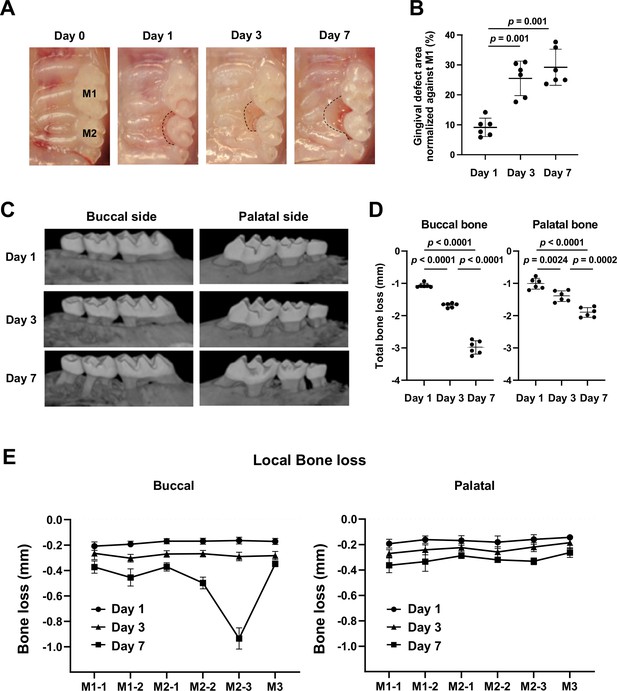
Gingival defect formation and alveolar bone loss in the ligature-induced periodontitis model in mice.
(A) A ligature (5.0 silk suture) was placed around the maxillary second molar (M2) of wild-type (WT) mice. Representative intra-oral photographs of the maxilla on day 0, prior to ligature placement (healthy gingiva), and on days 1, 3, and 7 after ligature placement. (B) The gingival defect area was measured and normalized to the circumferential area of the maxillary first molar (M1) (n = 6). Gingival defects appeared on day 3. (C) Representative micro-computed tomography (microCT) images of the maxilla taken from the lateral view. (D) Alveolar bone loss was determined from the total distance between the cementoenamel junction (CEJ) and the alveolar bone crest (ABC) of the buccal or palatal bone at six sites in the ligated side (n = 6). Alveolar bone loss was apparent on day 7. (E) Alveolar bone loss was assessed at the mesiobuccal cusp (M1-1), distobuccal cusp (M1-2), and distal cusp (M1-3) of the first molar, the mesiobuccal cusp (M2-1) and distobuccal cusp (M2-2) of the second molar, and the buccal cusp (M3) of the third molar by measuring the distance from the CEJ to the ABC on the buccal or palatal side of the alveolar bone (n = 6). Significance was determined by one-way ANOVA, with Tukey’s multiple-comparison test. Data are presented as mean values ± standard deviation (SD); p<0.05 was considered significant.

Identification of major cell types in mouse gingival tissue during periodontitis development by single-cell RNA sequencing (scRNA-seq).
Violin plots showing expression levels of cell-type marker genes in each major cell type: epithelial cells, cadherin 1 (Cdh1) and type XVII collagen (Col17a1); fibroblasts, type I collagen (Col1a1) and lumican (Lum); endothelial cells, selectin P (Selp) and selectin E (Sele); B cells, membrane spanning 4 domains A1 (Ms4a1) and cluster of differentiation 79A (Cd79a); T cells, epsilon subunit of T cell receptor complex (Cd3e) and cluster of differentiation 5 (Cd5); and myeloid cells, lysozyme 2 (Lyz2) and integrin subunit alpha M (Itgam).
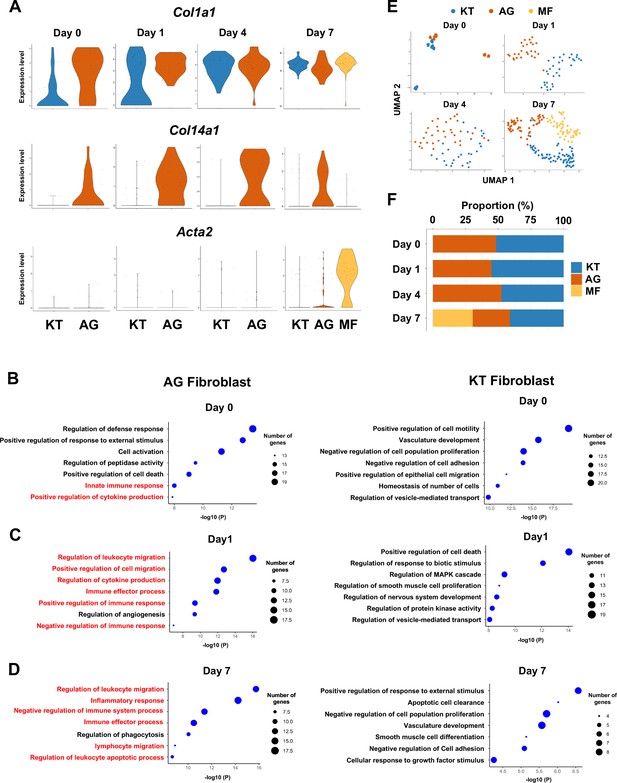
Fibroblasts activated to guide leukocyte migration (AG fibroblasts) are one of three fibroblast subpopulations in gingival tissue during periodontitis development.
(A) Violin plots showing gene expression levels of type I collagen (Col1a1), type XIV collagen (Col14a1), and smooth muscle aortic actin 2 (Acta2) in gingival fibroblast subpopulations during periodontitis development. AG, AG fibroblasts; KT, ‘keeping typical phenotype’ fibroblasts; MF, myofibroblasts. Gene Ontology (GO) enrichment analysis of the biological functions of AG fibroblasts and KT fibroblasts on day 0 without ligature placement (B) and on day 1 (C) and day 7 (D) after ligature placement. Gene clusters related to immune regulation (red) were identified in AG fibroblasts, and these clusters dominate after ligature placement. (E) t-distributed stochastic neighbor embedding (t-SNE) projection plots showing fibroblast subpopulations in gingival tissue during periodontitis development. Colors indicate cell type as follows: blue, KT fibroblasts; red, AG fibroblasts; and yellow, MFs. (F) Proportion plots showing the relative amounts of each fibroblast subpopulation on days 0, 1, 4, and 7.
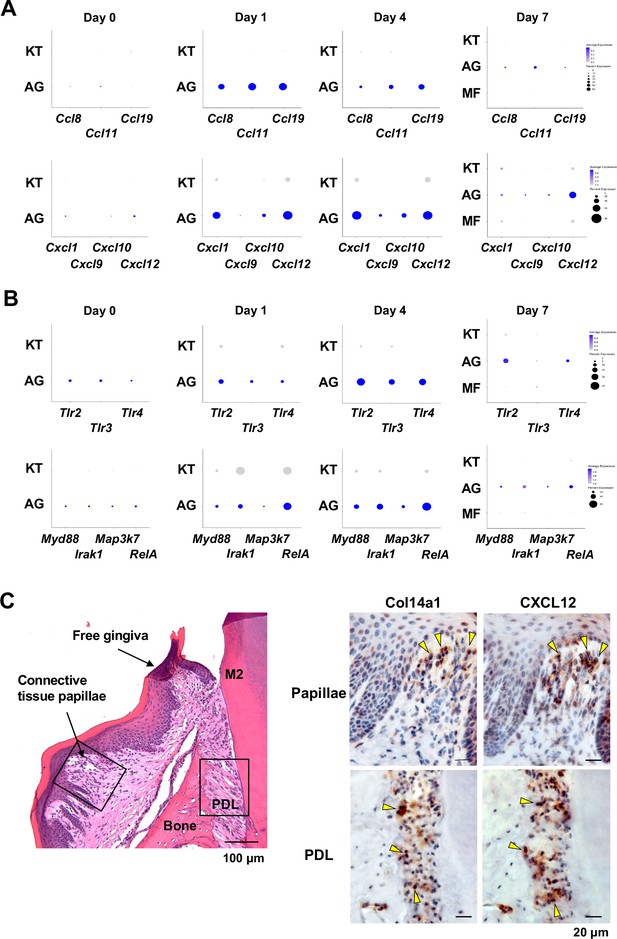
AG fibroblasts and immune surveillance in periodontitis development.
(A) Dot plots depicting expression levels of the CCL genes Ccl8, Ccl11, Ccl19, Cxcl1, Cxcl9, Cxcl10, and Cxcl12 in gingival fibroblast subpopulations during periodontitis development. (B) Dot plots depicting expression levels of the Toll-like receptor (TLR) and related genes Tlr2, Tlr3, Tlr4, Myd88, Irak1, Map3k7, and Rela in gingival fibroblast subpopulations during periodontitis development. Upregulation of chemokines and TLR-related molecules is predominantly observed in the AG fibroblast subpopulation. (C) Hematoxylin and eosin (HE) staining and immunohistochemical (IHC) staining for COL14A1 and CXCL12 in periodontal tissue on day 1; scale bars, 100 µm (HE) and 20 µm, (IHC). Yellow arrows indicate COL14A1- and CXCL12-positive cells in the connective tissue papillae and periodontal ligament (PDL).
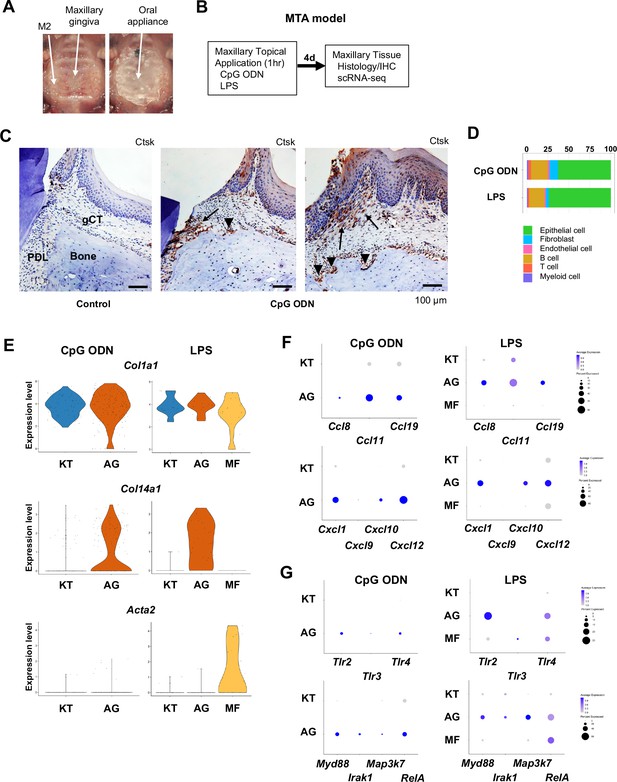
Characterization of AG fibroblasts in the discrete maxillary topical application (MTA) model using cytidine phosphate guanosine oligonucleotide (CpG ODN) (TLR9 ligand) and LPS (TLR2/4 ligand).
(A) The MTA model was developed to discretely test the selected oral microbial pathogens. The selected pathogen was topically applied directly on the maxillary gingiva between the molars and covered by a custom fabricated oral appliance for 1 hr. (B) In this study, microbial DNA TLR9 ligand, unmethylated CpG ODN, and TLR2/4 ligand P. gingivalis lipopolysaccharide (LPS) were selected in the MTA model. After 1 hr exposure, mice were returned to the vivarium for 4 d and the maxillary tissue was harvested for histology or scRNA-seq. (C) The MTA of CpG ODN developed localized periodontal ligament (PDL) and gingiva connective tissue (gCT) degradation evidenced by the expression of cathepsin K (Ctsk; arrows). There were signs of localized bone resorption by Ctsk+ osteoclasts (arrowheads) on the surface of alveolar bone (Bone). (D) Gingival cell composition by scRNA-seq of the MTA of CpG ODN or LPS revealed early stage of gingival inflammation, equivalent to day 1 of the ligature-induced periodontitis model. (E) The fibroblastic gene expression signature revealed the presence of KT and AG fibroblasts by the MTA of CpG ODN, whereas the MTA of LPS induced myofibroblast (MF). (F) AG fibroblasts of the MTA of CpG ODN and LPS were activated to express CCL and CXCL chemokines. (G) The MTA of CpG ODN did not upregulate Tlr9, whereas the MTA of LPS increased the expression of Tlr2/4 in AG fibroblasts. However, the both the MTA of CpG ODN and LPS increased the expression of TLR downstream molecules.
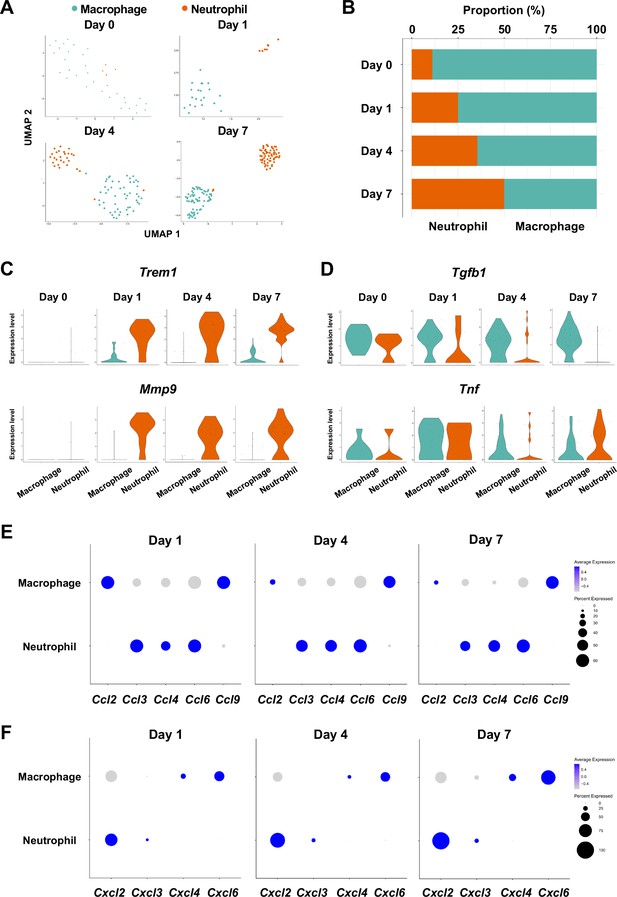
Myeloid cell composition and activity in gingival tissue during in periodontitis development.
(A) t-distributed stochastic neighbor embedding (t-SNE) projection plots showing myeloid cell subpopulations in gingival tissue during periodontitis development on days 0, 1, 4, and 7. Colors indicate cell type as follows: green, macrophages; and red, neutrophils. (B) Proportion plots showing the relative amounts of neutrophils and macrophages on days 0, 1, 4, and 7. (C) Violin plots showing Trem1 and Mmp9 expression levels in myeloid cells on days 0, 1, 4, and 7; both genes are upregulated in neutrophils after ligature placement. (D) Violin plots showing Tgfb1 and Tnf expression in myeloid cells on days 0, 1, 4, and 7; no obvious induction is observed in response to ligature placement. (E) Dot plots depicting expression levels of the C motif chemokine ligand (CCL) genes Ccl2, Ccl3, Ccl4, Ccl6, Ccl9. (F) The expression of CXC motif chemokine ligand (CXCL) genes Cxcl2, Cxcl3, Cxcl4, and Cxcl9. Chemokine expression in myeloid cells was unrelated to progression of gingival inflammation from days 1–7.
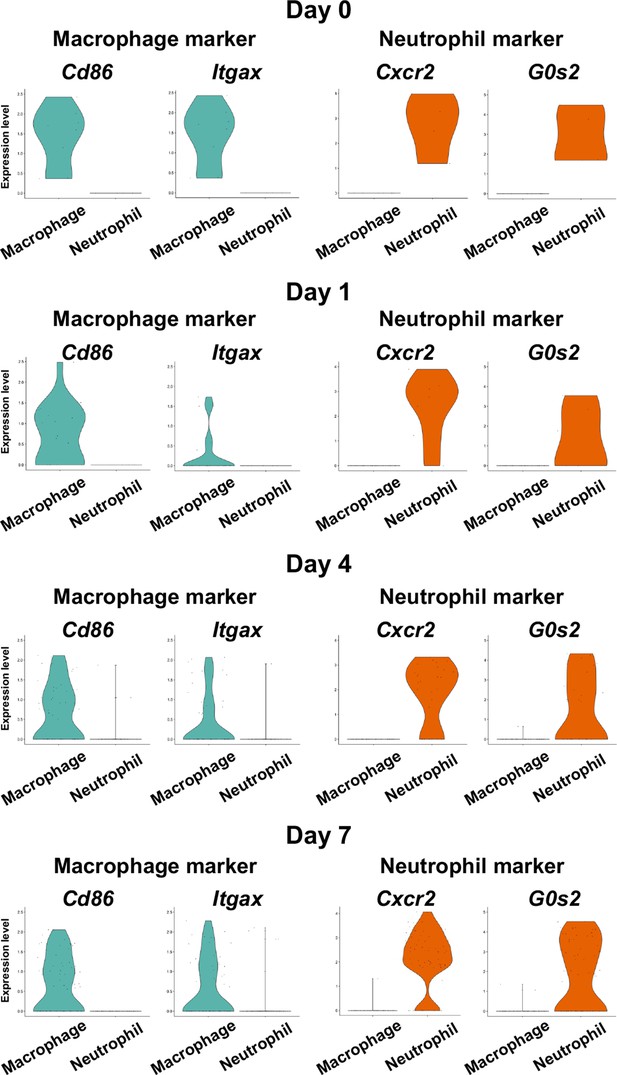
Identification of myeloid cell subpopulations in mouse gingival tissue during periodontitis development by scRNA-seq.
Violin plots showing expression levels of macrophage and neutrophil marker genes in myeloid cell subpopulations: macrophages, cluster of differentiation 86 (Cd86), and integrin subunit alpha X (Itgax); and neutrophils, CXC motif chemokine receptor 2 (Cxcr2), and G0/G1 switch gene 2 (G0s2).
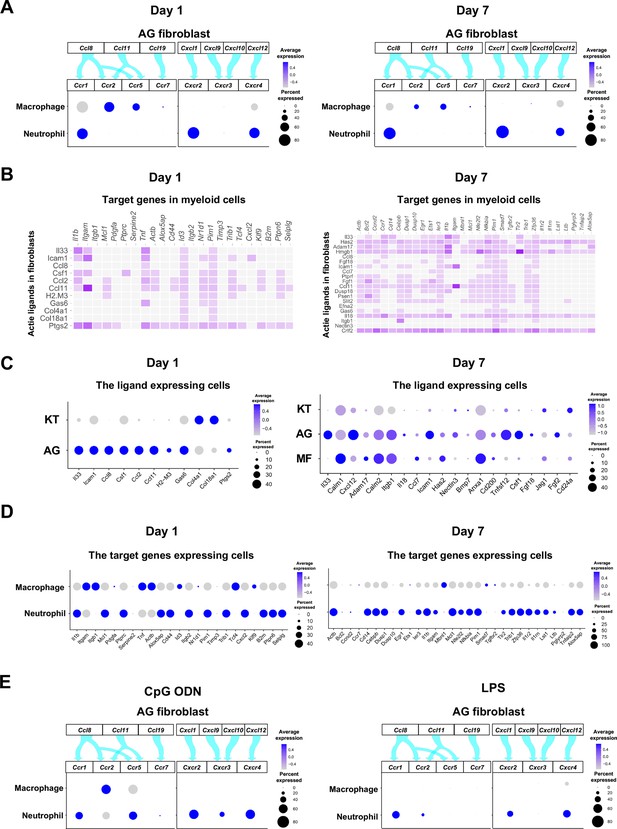
Role of AG fibroblasts in myeloid cell activation.
(A) Interaction between chemokine ligands expressed by AG fibroblasts and their putative chemokine receptors expressed by myeloid cells during periodontitis development. Dot plots depicting expression levels of the CC chemokine receptor (CCR) and the CXC chemokine receptor (CXCR) genes Ccr1, Ccr2, Ccr5, Ccr7, Cxcr2, Cxcr3, and Cxcr4 in myeloid cell subpopulations on days 1 and 7 following ligature placement. (B) NicheNet ligand–target matrix indicating the regulatory potential between active ligands expressed in fibroblasts and target genes expressed in myeloid cells from the p-EMT program on days 1 and 7. (E) Dot plot depicting expression levels of active ligand genes from panel (B) in fibroblast subpopulations on days 1 and 7. (D) Dot plot depicting expression levels of target genes from panel (B) in myeloid cell subpopulations on days 1 and 7. Results suggest a strong intercellular communication network from AG fibroblasts to neutrophils. (E) Interaction between chemokine ligands expressed by AG fibroblasts and their putative chemokine receptors expressed by myeloid cells in the maxillary topical application (MTA) model of cytidine phosphate guanosine oligonucleotide (CpG ODN) and lipopolysaccharide (LPS).
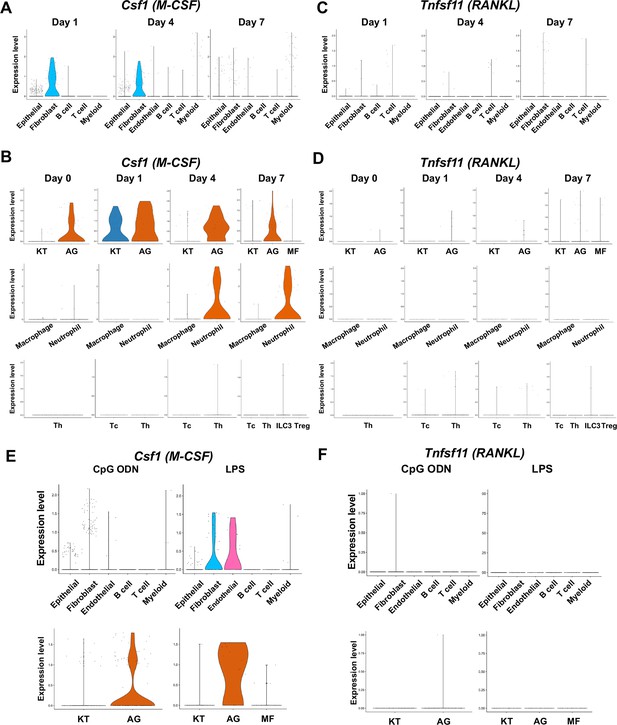
The cellular source of osteoclastogenic cytokines: macrophage-colony stimulating factor (M-CSF) and receptor activator of nuclear factor kappa-Β ligand (RANKL) in the ligature-induced model and the maxillary topical application (MTA) model.
(A) Violin plots showing expression levels of the M-CSF-encoding gene, Csf1 (M-CSF), in each major cell type from the ligature-induced periodontitis model. (B) Violin plots showing expression levels of Csf1 in fibroblast subpopulations, myeloid cell subpopulations, and T cell subpopulations. (C) Violin plots showing expression levels of the RANKL-encoding gene, Tnfsf11, in each major cell type. (D) Violin plots showing expression levels of Tnfsf11 in fibroblast subpopulations, myeloid cell subpopulations, and T cell subpopulations. (E) Violin plots of Csf1-expressing cells in the MTA model. (F) Violin plots of Tnfsf11-expressing cells in the MTA model. AG fibroblasts predominantly expressed Tnfsf11.
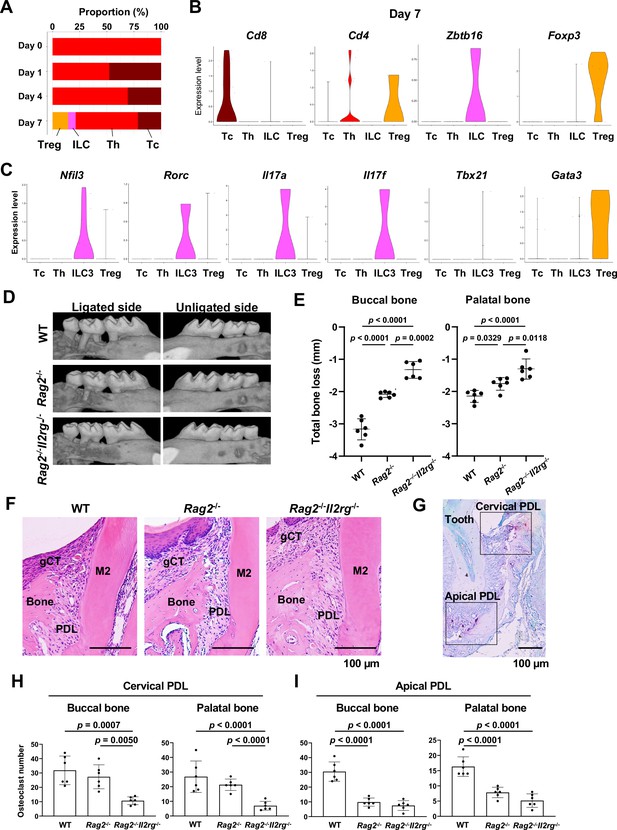
Type 3 innate lymphoid cells (ILC3s) are critical for cervical alveolar bone resorption in the ligature-induced periodontitis development.
(A) Proportion plots showing the relative amounts of T cell subpopulations in gingival tissue during periodontitis development. Treg, T regulatory cells; ILC, innate lymphoid cells; Th, T helper cells; Tc, cytotoxic T cells. (B) Violin plots showing expression levels of the T cell marker genes Cd8 (Tc), Cd4 (Th), Zbtb16 (ILC), and Foxp3 (Treg) on day 7 following ligature placement. (C) Violin plots showing expression levels of Nfil3, Rorγ, Il17a, Il17f, Tbx21, and Gata3 on day 7 following ligature placement. These gene signatures indicate that gingival ILCs primarily comprise ILC3s. (D) Representative micro-computed tomography (microCT) images of the maxilla taken from the lateral view for the ligated side and from the contralateral view for the unligated side. (E) Alveolar bone loss was determined from the total distance between the cementoenamel junction (CEJ) and the alveolar bone crest (ABC) of the buccal bone or palatal bone at six sites in the ligated side (n = 6). (F) HE staining of the periodontal tissue on day 7. gCT, gingival connective tissue; Bone, alveolar bone; PDL, periodontal ligament; scale bars, 100 µm. (G) Tartrate-resistant acid phosphatase (TRAP) staining of periodontal tissue from WT mice on day 7; scale bar, 100 µm. Total number of TRAP-positive cells in a 0.01 mm2 area of the buccal and palatal bone in the cervical PDL site (H) and apical PDL site (I) (n = 6). Significance was determined by ANOVA, with Tukey’s multiple-comparison test. Data are presented as mean values ± SD; p<0.05 was considered significant.
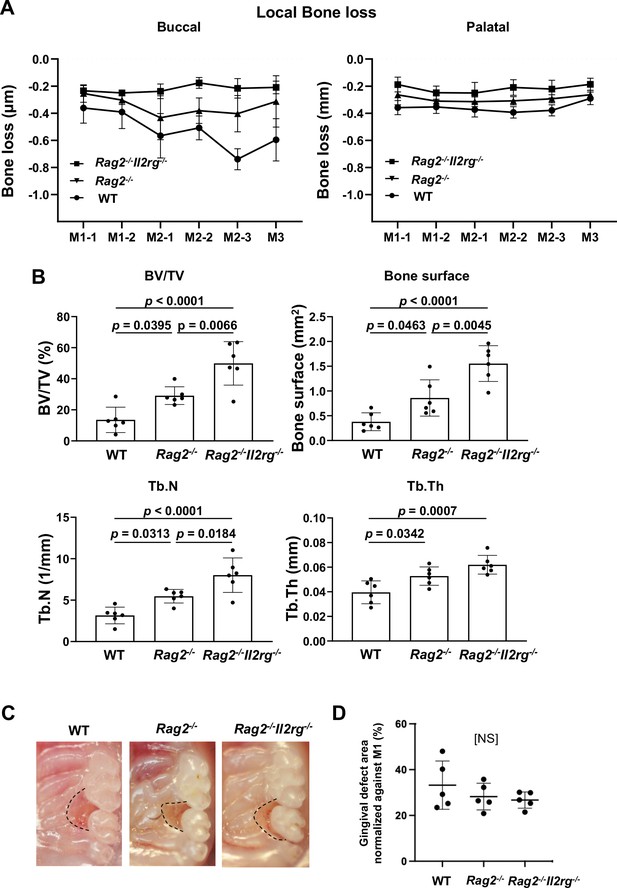
Effects of innate lymphoid cell (ILC) deletion on alveolar bone loss in the mouse periodontitis model.
(A) Alveolar bone loss was assessed at six sites by measuring the distance from the cementoenamel junction (CEJ) to the alveolar bone crest (ABC) on the buccal or palatal side of the alveolar bone of wild-type (WT), Rag2-/-, and Rag2-/-Il2rg-/- mice on day 7 following ligature placement (n = 6). (B) Bone volume/total volume (BV/TV), bone surface, trabecular number (Tb.N), and trabecular thickness (Tb.Th) in the buccal side of alveolar bone of the second molar were measured on day 7 (n = 6). (C) Representative intra-oral photographs of maxilla from WT, Rag2-/-, and Rag2-/-Il2rg-/- mice taken on day 7 following ligature placement. (D) The gingival defect area was measured and normalized to the circumferential area of M1 (n = 5). Significance was determined by ANOVA, with Tukey’s multiple-comparison test. Data are presented as mean values ± SD; p<0.05 was considered significant.

Human periodontitis phenotype.
(A) Panoramic radiograph of a human patient diagnosed with generalized periodontitis. The alveolar bone resorption was only noted at the cervical alveolar bone (arrows), whereas the apical alveolar bone was well maintained and not affected by periodontitis. (B) Periapical dental radiograph and the clinical pictures during the periodontal surgery. A localized alveolar bone loss (arrows) was noted at the cervical periodontal ligament area.
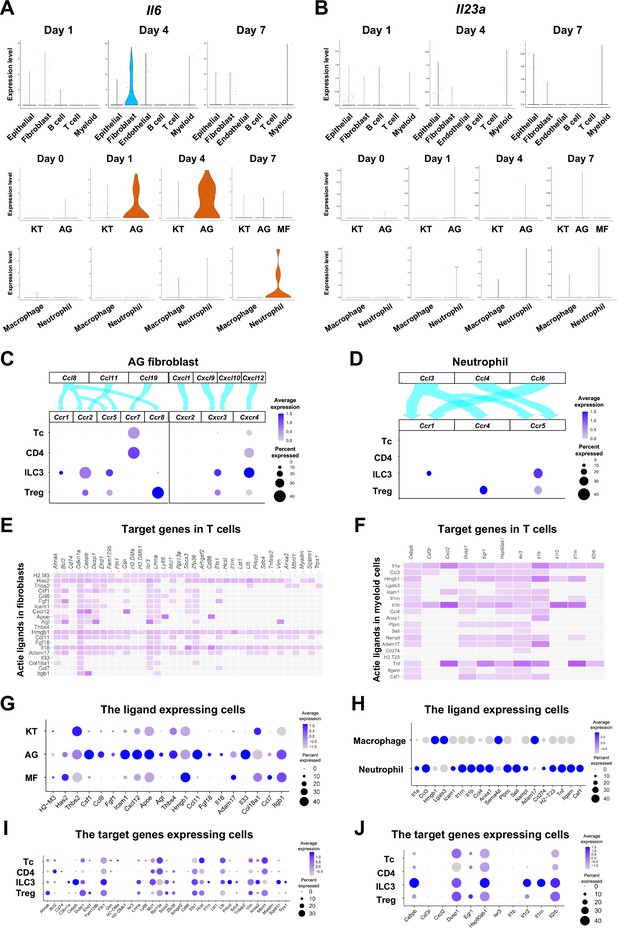
The role of AG fibroblasts and neutrophils in type 3 innate lymphoid cell (ILC3) development in periodontitis.
Violin plots showing expression levels of the genes encoding interleukin (IL)-6 (Il6) (A) and IL-23 (Il23a) (B) in each major cell type, fibroblast subpopulations, and myeloid cell subpopulations during periodontitis development. (C) Interaction between chemokine ligands strongly expressed by AG fibroblasts and their putative chemokine receptors expressed by T cells, including ILCs. Dot plots depict gene expression levels of Ccr1, Ccr2, Ccr5, Ccr7, Ccr8, Cxcr2, Cxcr3, and Cxcr4 in T cell subpopulations on day 7 following ligature placement. (D) Interaction between chemokine ligands strongly expressed by neutrophils and their putative chemokine receptors expressed by T cells. Dot plots depicting gene expression levels of Ccr1, Ccr4, and Ccr5 in T cell subpopulations on day 7 following ligature placement. (E) NicheNet ligand–target matrix denoting the regulatory potential between active ligands in fibroblasts and target genes in T cells from the p-EMT program on day 7 following ligature placement. (F) NicheNet ligand–target matrix denoting the regulatory potential between active ligands in myeloid cells and target genes in T cells from the p-EMT program on day 7 following ligature placement. (G) Dot plot depicting expression levels of active ligand genes from panel (E) in fibroblast subpopulations. (H) Dot plot depicting expression levels of active ligand genes from panel (F) in myeloid cell subpopulations. (I) Dot plot depicting expression levels of target genes from pane (E) in T cell subpopulations. (J) Dot plot depicting expression levels of target genes from panel (F) in T cell subpopulations.

Epithelial cell subpopulations involved in periodontitis development in mice.
(A) t-distributed stochastic neighbor embedding (t-SNE) projection plots showing epithelial cell subpopulations in gingival tissue on days 1, 4, and 7 following ligature placement. Colors indicate cell type as follows: dark green, epithelial cell population 1 (Epi 1); sea green, epithelial cell population 2 (Epi 2); pale green, epithelial cell population 3 (Epi 3); light green, epithelial cell population 4 (Epi 4); and light blue, epithelial cell population expressing epithelial–mesenchymal transition (EMT) genes. (B) Heatmap showing expression of the top-10 differentially expressed genes in Epi 1, Epi 2, Epi 3, Epi4, and EMT. (C) Violin plots showing gene expression levels of keratin 5 (Krt5), keratin 14 (Krt14), cadherin 11 (Cdh11), Col1a1, tenascin C (Tnc), and smooth muscle aortic actin 2 (Acta2) in epithelial cell subpopulations. (D) Violin plots showing gene expression levels of interleukin 6 (Il6) and interleukin 23A (Il23a) in epithelial cell subpopulations.
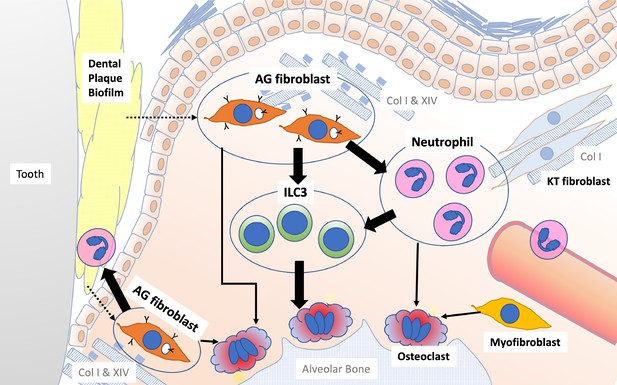
Schematic overview of the newly proposed AG fibroblast–neutrophil–ILC3 axis.
We propose that periodontal inflammation is initiated by the activation of AG fibroblasts, which secrete chemokines and cytokines that recruit neutrophils to sites of tissue damage. Activated neutrophils and AG fibroblasts, in turn, activate ILC3s, leading to the production of proinflammatory IL-17 cytokines. Ultimately, cervical alveolar bone resorption is facilitated by a localized osteoclastogenic environment, induced by activated ILC3s, together with AG fibroblasts, neutrophils, myofibroblasts, and gingival epithelial cells, including those with an epithelial–mesenchymal transition (EMT) phenotype.





