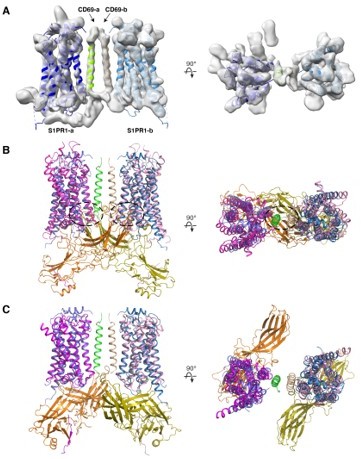
Transmembrane protein CD69 acts as an S1PR1 agonist
Figures
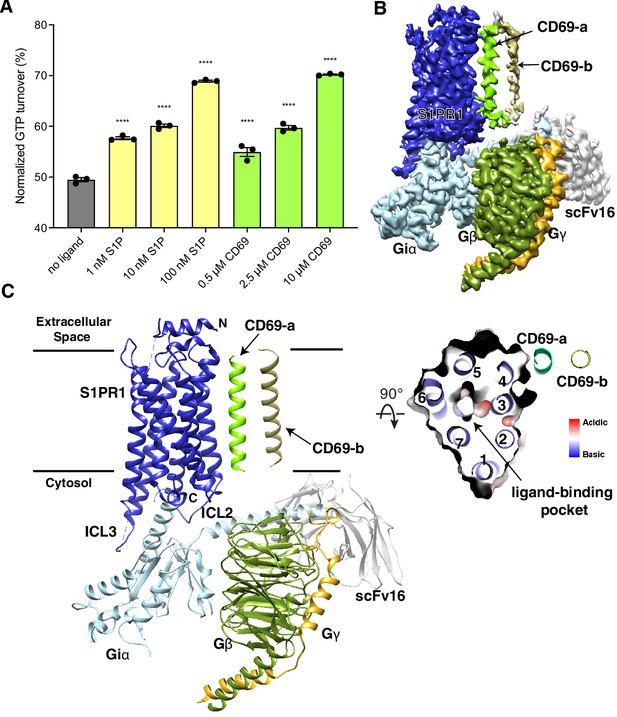
Overall structure of human CD69-S1PR1-Gi complex.
(A) S1PR1-induced GTP turnover for Gi1 in the presence of purified CD69 or S1P. Luminescence signals were normalized relative to the condition with Gi1 only. Data are mean ± s.e.m. of three independent experiments. One-way ANOVA with Tukey’s test; ****p<0.0001. Experiments were repeated at least three times with similar results. (B) Cryo-EM map of human CD69 bound S1PR1-Gi complex. (C) Cartoon presentation of the complex in the same view and color scheme as shown in (B). Slab view of S1PR1 from the extracellular side showing that the orthosteric binding pocket is vacant.
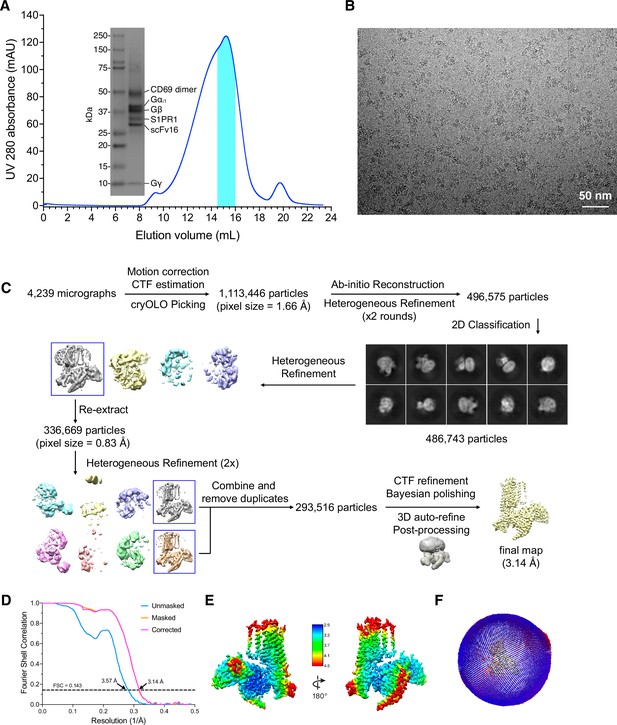
Cryo-EM reconstruction of CD69 bound S1PR1-Gi complex.
(A) Size exclusion column profile of CD69-S1PR1-Gi complex. Elution fractions of the peak indicated by cyan color were pooled and concentrated for use in grid vitrification. (B) Representative micrograph after motion correction. (C) Data processing workflow of CD69-S1PR1-Gi complex. Representative 2D classes are shown. (D) FSC curve of the final reconstruction in Post-processing of Relion. (E) Local resolution display of the reconstructed map. Local resolution was estimated within Relion. (F) Angular distribution of particles used in the final reconstruction of the complex.
-
Figure 1—figure supplement 1—source data 1
Original uncropped SDS-PAGE gels for data in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/88204/elife-88204-fig1-figsupp1-data1-v2.zip
-
Figure 1—figure supplement 1—source data 2
Uncropped SDS-PAGE gels for data in Figure 1—figure supplement 1 with the relevant bands labeled.
- https://cdn.elifesciences.org/articles/88204/elife-88204-fig1-figsupp1-data2-v2.pdf
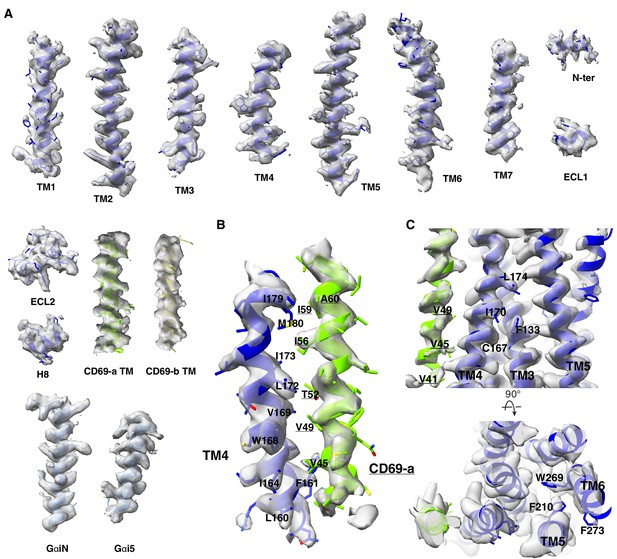
The cryo-EM density map of CD69-bound S1PR1-Gi complex.
(A) Representative regions of CD69, S1PR1, and Gαi1 are shown. (B) The cryo-EM density maps of the CD69 residues. (C) The cryo-EM density maps of the S1PR1 residues. The density map is drawn by ChimeraX.
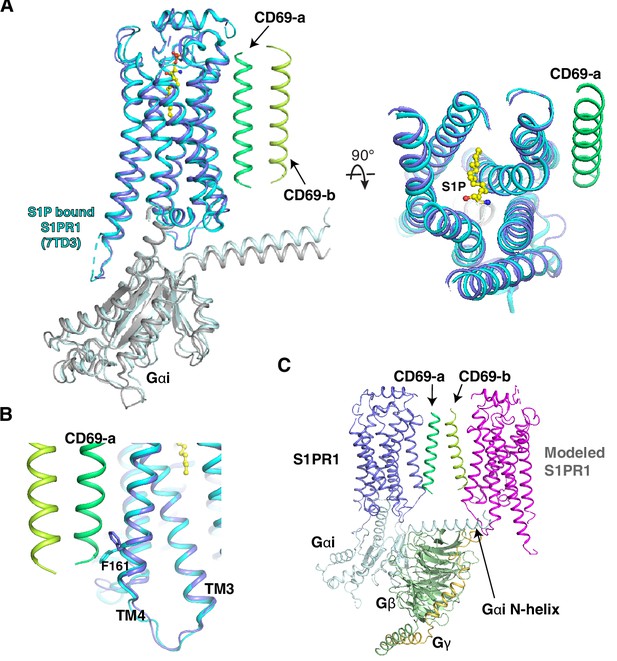
Structural comparison between CD69-bound S1PR1 and S1P-bound S1PR1.
(A) Structures of S1PR1 binding with CD69 and S1P. CD69-bound receptor in blue and S1P-bound S1PR1 (PDB code: 7TD3) in cyan. S1P is shown as balls and sticks in yellow. (B) The flipping of F1614.43 of the structures. (C) A model of 2:2 CD69-S1PR1 complex with Gi heterotrimer. The steric clash between the modeled S1PR1 and the N-helix of Gai is indicated by an arrow.
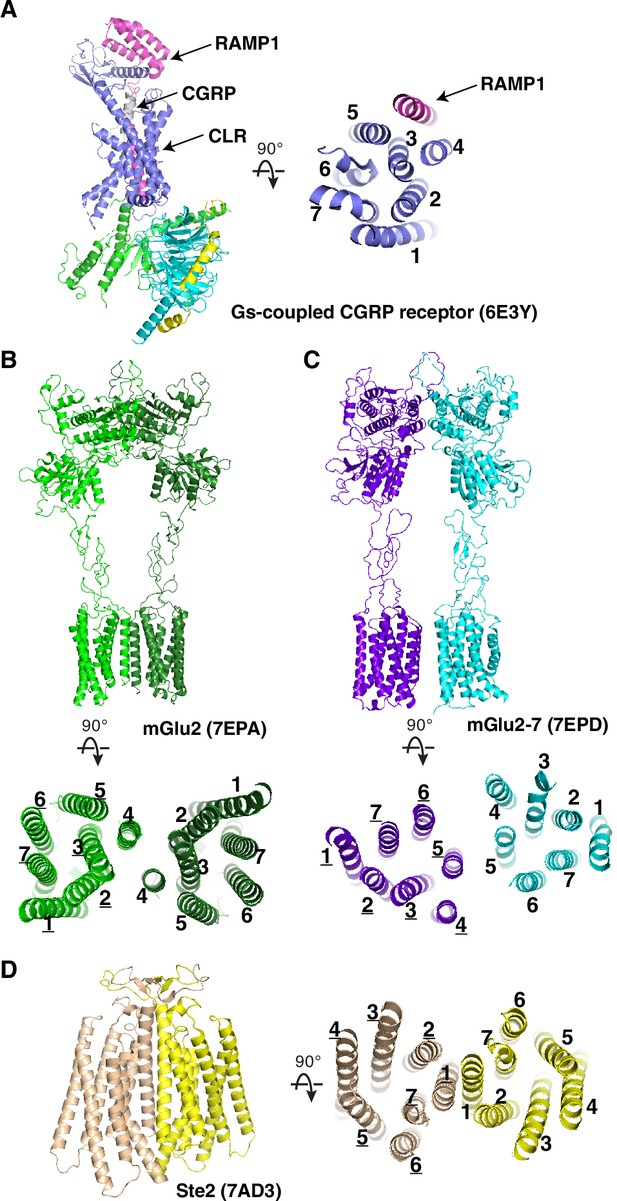
Structures of homodimeric and heterodimeric GPCRs.
(A) Overall structures of Gs-protein-coupled CGRP receptor with CGRP. (B) Overall structures of mGlu2 homodimer. (C) mGlu2–mGlu7 heterodimer. (D) the class D GPCR Ste2. Transmembrane helices are labeled and PDB codes are shown in parentheses.
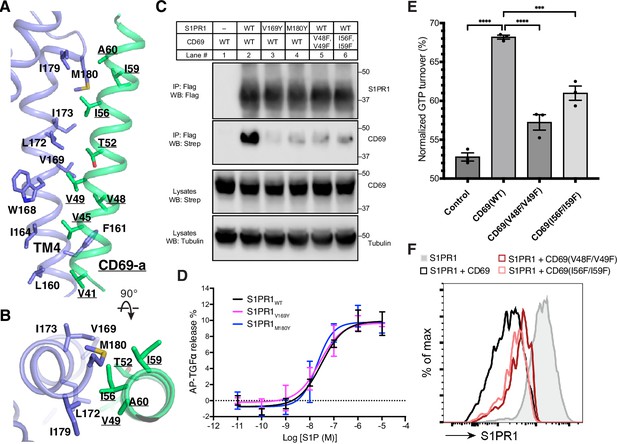
The binding interface between CD69 and S1PR1.
(A) and (B) Detailed interactions between CD69-a and TM4 of S1PR1. Residues that contribute to complex formation are labeled. CD69 is shown in green and S1PR1 in slate. (C) S1PR1-Flag and CD69-StrepII co-immunoprecipitation assay in transfected HEK293 GnTI- cells from one experiment that is representative of three. (D) Dose-response curves of S1PR1WT, S1PR1V169Y and S1PR1M180Y for the TGFα shedding assay using S1P. Data are mean ± s.d. (n=3). (E) S1PR1-induced GTP turnover for Gi1 in the presence of purified wild-type and mutant CD69. Luminescence signals were normalized relative to the condition with Gi1 only. Data are mean ± s.e.m. of three independent experiments. One-way ANOVA with Tukey’s test; ***p<0.001, ****p<0.0001. Experiments in (C)-(E) were repeated at least twice with similar results. (F) Flow cytometric analysis of S1PR1 surface expression on WEHI231 lymphoma cells transduced with S1PR1 and CD69 wild-type and mutant constructs as indicated. From one experiment that is representative of three.
-
Figure 2—source data 1
Original uncropped western blots for data in Figure 2.
- https://cdn.elifesciences.org/articles/88204/elife-88204-fig2-data1-v2.zip
-
Figure 2—source data 2
Uncropped western blots for data in Figure 2 with the relevant bands labeled.
- https://cdn.elifesciences.org/articles/88204/elife-88204-fig2-data2-v2.pdf
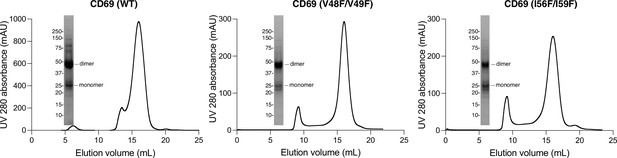
Size exclusion column profiles of CD69 wild type and mutants.
CD69 is a dimer in size exclusion buffer, but partially dissociated to be monomer when resolved in SDS-PAGE gel.
-
Figure 2—figure supplement 1—source data 1
Original uncropped SDS-PAGE gels for data in Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/88204/elife-88204-fig2-figsupp1-data1-v2.zip
-
Figure 2—figure supplement 1—source data 2
Uncropped SDS-PAGE gels for data in Figure 2—figure supplement 1 with the relevant bands labeled.
- https://cdn.elifesciences.org/articles/88204/elife-88204-fig2-figsupp1-data2-v2.pdf
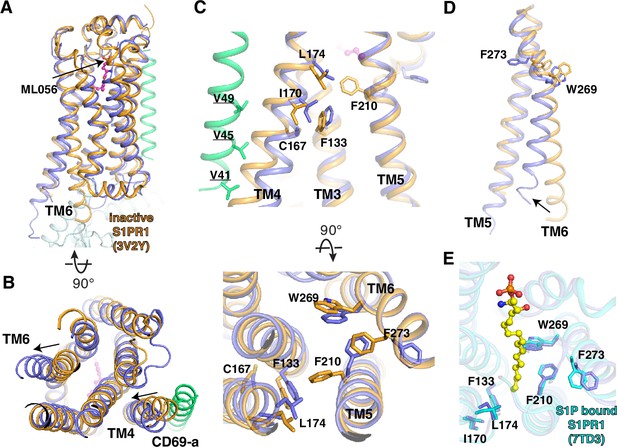
Comparison between CD69-bound S1PR1 and ML056- or S1P-bound S1PR1.
(A) Overall structures of S1PR1 binding with CD69 and ML056. The CD69-bound S1PR1 structure was aligned to ML056-bound inactive S1PR1 (PDB code: 3V2Y). ML056-bound receptor is shown in brown, CD69-bound receptor in blue, and the TM of CD69 in green. The same color scheme is used (C) and (D). (B) The movements of TM4 and TM6 of CD69-bound S1PR1 compared with ML056-bound inactive S1PR1. (C) Residues involved in the TM movements. (D) TM6 movement around W2696.48 and F2736.52. (E) Comparison between CD69-bound S1PR1 and S1P-bound S1PR1 (PDB code: 7TD3). Residues in the ligand binding pocket are shown. CD69-bound receptor in blue and S1P-bound S1PR1 in cyan. S1P is shown as balls and sticks in yellow.
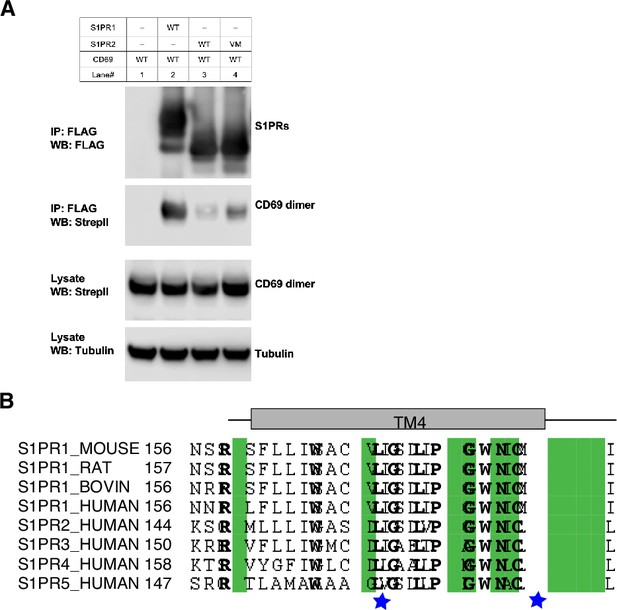
S1PR1 specificity for CD69 binding.
(A) Co-immunoprecipitation assay in transfected HEK293 GnTI- cells from one experiment that is representative of three. (B) Sequence alignment of S1PR homologues. The different residues among S1PRs which are crucial for CD69 binding are indicated by blue stars.
-
Figure 3—figure supplement 1—source data 1
Original uncropped western blots for data in Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/88204/elife-88204-fig3-figsupp1-data1-v2.zip
-
Figure 3—figure supplement 1—source data 2
Uncropped western blots for data in Figure 3—figure supplement 1 with the relevant bands labeled.
- https://cdn.elifesciences.org/articles/88204/elife-88204-fig3-figsupp1-data2-v2.pdf
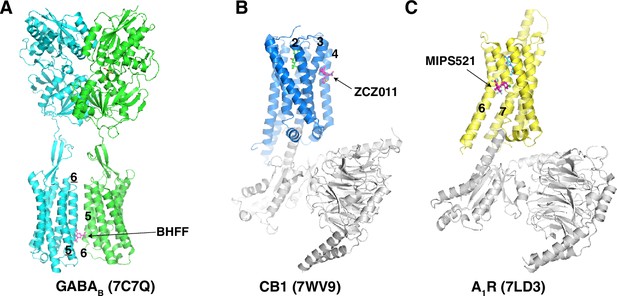
Structures of GPCRs with their positive allosteric modulators.
(A) Structure of GABAB. The BHFF, a positive allosteric modulator (PAM) of GABAB, is shown as magenta sticks. (B) Structure of CB1. The ZCZ011, a PAM of CB1, is shown as magenta sticks. (C) Structure of A1R. The MIPS521, a PAM of A1R, is shown as magenta sticks. Transmembrane helices for PAM binding are labeled and PDB codes are shown in parentheses.
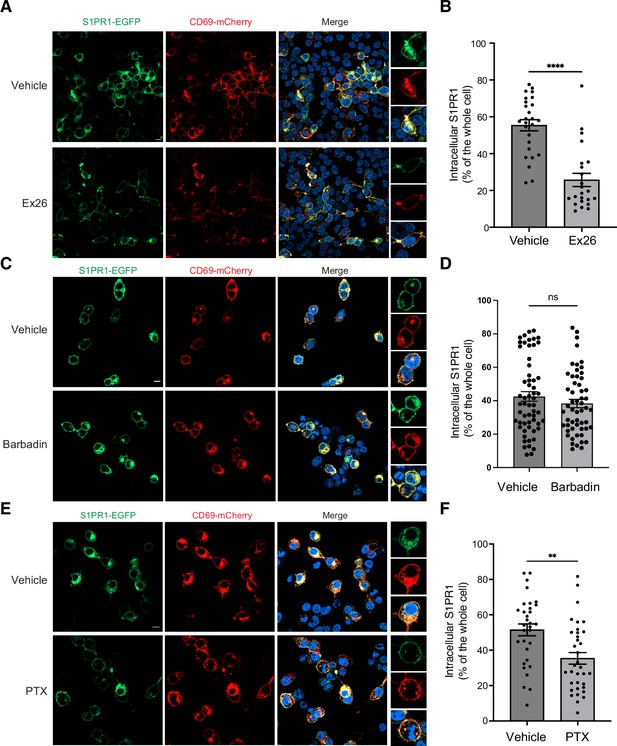
CD69 induced S1PR1 internalization.
(A) HEK293 cells were treated with 2 μM Ex26 or vehicle for 12 hr and imaged using confocal microscopy. Scale bar, 10 μm. (B) Quantification of intracellular S1PR1 of the cells in (A). (C) HEK293 cells were treated with 20 μM Barbadin for 12 hr and imaged for analysis. Scale bar, 10 μm. (D) Quantification of intracellular S1PR1 of the cells in (C). (E) HEK293 cells were treated with 200 ng/ml pertussis toxin (PTX) for 12 hr and imaged for analysis. Scale bar, 10 μm. (F) Quantification of intracellular S1PR1 of the cells in (E). Data are mean ± s.e.m. Two-sided Welch’s t-test; ns, not significant, **p<0.01, ****p<0.0001. All experiments were repeated at least three times with similar results.
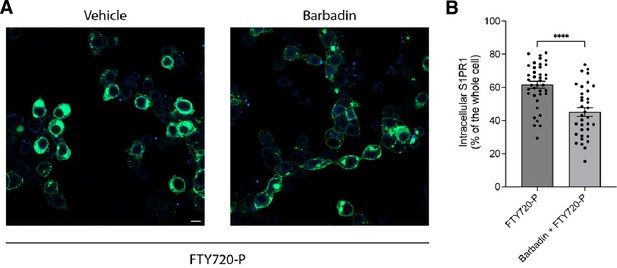
Barbadin alters the FTY720-P mediated S1PR1 internalization.
(A) HEK293 cells were treated with 20 mM Barbadin for 2 hr followed by vehicle- or FTY720-P-treatment for 1 hr and imaged using confocal microscopy. Scale bar, 10 mm. (B) Quantification of intracellular S1PR1 of the cells in (A). Data are mean ± s.e.m. Two-sided Welch’s t-test; ****p<0.0001. Experiments were repeated at least twice with similar results.
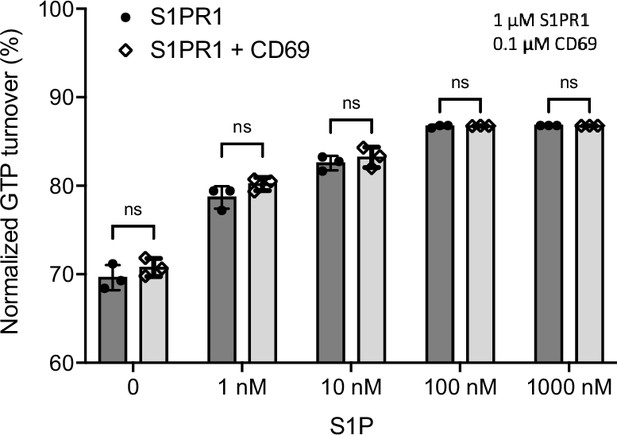
S1PR1-induced GTP turnover for Gi1 in the presence of purified CD69 and S1P.
Luminescence signals were normalized relative to the condition with Gi1 only. Data are mean ± s.e.m. of three independent experiments. Two-sided Student’s t-test; ns, not significant. Experiments were repeated at least twice with similar results.
Tables
Cryo-EM data collection, processing, and refinement statistics.
| Structure | CD69-S1PR1-Gi-scFv16 |
|---|---|
| PDB | 8G94 |
| EMDB | EMD-29861 |
| Data collection/ processing | |
| Magnification | 105,000 |
| Voltage (kV) | 300 |
| Pixel size (Å) | 0.83 |
| Defocus range (μm) | 1.0–2.0 |
| Electron exposure (e-/Å2) | 60 |
| Symmetry imposed | C1 |
| Initial particles (No.) | ~1.1 million |
| Final particles (No.) | 293,516 |
| Map resolution (Å) | 3.14 |
| FSC threshold | 0.143 |
| Map resolution range (Å) | 25–3.0 |
| Refinement | |
| Model Resolution (Å) | 3.3 |
| FSC threshold | 0.5 |
| Map sharpening B-factor (Å2) | –60 |
| Model composition | |
| Non-hydrogen atoms | 9223 |
| Protein residues | 1187 |
| Ligand | 0 |
| B-factors (Å2) | |
| Protein | 98.23 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 0.702 |
| Validation | |
| MolProbity score | 1.64 |
| Clashscore | 6.49 |
| Rotamers outliers (%) | 0.00 |
| Ramachandran plot (%) | |
| Favored | 95.87 |
| Allowed | 4.13 |
| Outliers | 0.00 |