Water and chloride as allosteric inhibitors in WNK kinase osmosensing
Figures
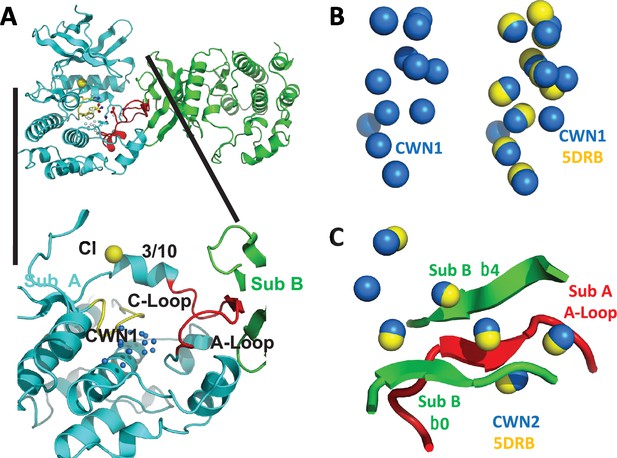
Conserved water networks in WNK1/SA.
(A) Location of conserved water network 1 (CWN1) in Subunit A of uWNK1/SA (PDB file 6CN9) shown as a closeup from the uWNK/SA dimer. CWN1 in marine, 14 water molecules. Subunit A, cyan, Subunit B, green, Activation Loop, red, and Catalytic Loop, yellow. (B) Conservation of waters in two crystal structures of uWNK1/SA, PDB 6CN9 (waters marine) and 5DRB (waters yellow). (C) Conserved water network 2 (CWN2) is in the subunit interface (same coloring as in (A)). N-terminal domains are superimposed.
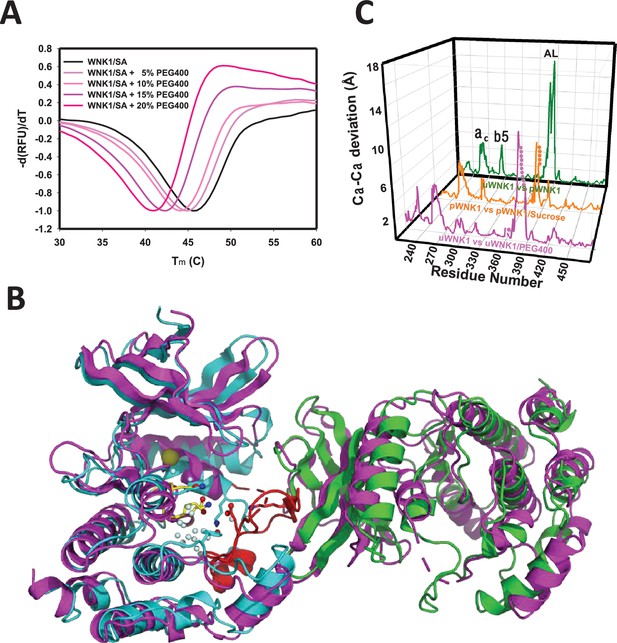
Effects of PEG400 on the WNK1/SA dimer and space group.
(A) Differential scanning fluorimetry of WNK1 SA in increasing PEG400 (showing destabilization). (B) WNK1/SA/PEG400 (PDB 9D3F) has a single subunit in the asymmetric unit (magenta), shown with a symmetry mate (magenta) and overlayed with the asymmetric dimer of uWNK1/SA (PDB 6CN9, shown in cyan and green as in Figure 1). (C) Osmolyte-induced conformational changes as a function of sequence. WNK1/SA (PDB file 6CN9) versus WNK1/SA/PEG400, pink trace, pWNK1 (PDB 5W7T) versus pWNK1/sucrose (Akella et al., 2020), orange trace, and uWNK1 (6CN9) pWNK1 (5W7T), green trace.
-
Figure 2—source data 1
Differential scanning fluorimetry of WNK1/SA in PEG400 showing destabilization.
- https://cdn.elifesciences.org/articles/88224/elife-88224-fig2-data1-v1.xlsx
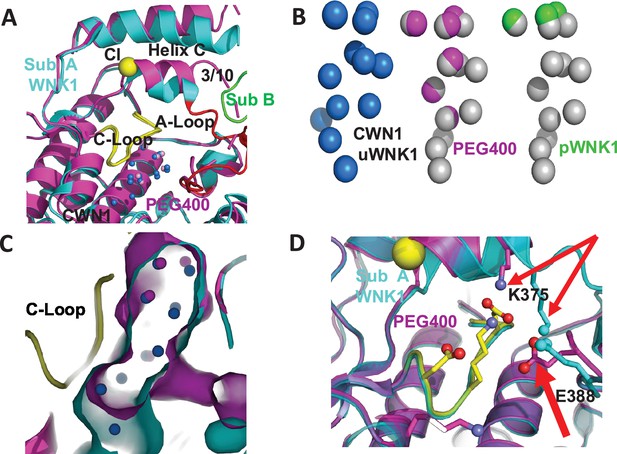
PEG400-induced conformational change in the active site of WNK1/SA.
(A) PEG400-induced conformational changes in WNK1/SA. WNK1/SA Sub A (cyan), Sub B (green), superimposed with WNK1/SA/PEG400 (magenta). Conformational changes occur in the active site in both the 3/10 chloride-binding helix and the Activation Loop. (B) Comparison of conserved water network 1 (CWN1) in uWNK1 (blue), PEG400 (magenta), and in pWNK (lime). (C) Surface rendering in WNK1/SA/PEG400 showing the reduction in space near CWN1. (D) Closeup of the Activation Loop highlighting residues E388 and K375 that move significantly in PEG400 (red arrows), affecting the 3/10 helix. Diagrams made in PyMOL.
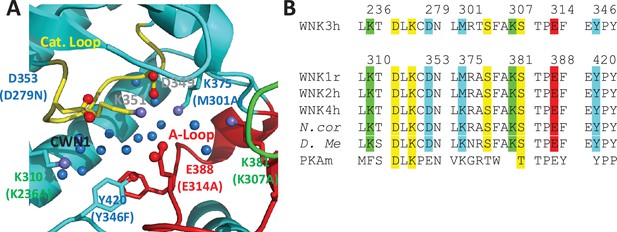
Residues in the AL-CL Cluster and positions mutated.
(A) AL-CL Cluster in uWNK1 (PDB file 6CN9). Labels in WNK1 numbering (with WNK3 numbers in parenthesis). Cartoon coloring is the same as Figure 1A. Pan-kinase-conserved catalytic residues (D349 and K351) are labeled in gray. AL-CL residue labels are colored to indicate mutant assay results: mutants more active than wild-type, red, similar to wild-type, green, and less active than wild-type, blue. (B) Sequence conservation in the AL-CL Cluster and neighboring residues. Pan-kinase-conserved Catalytic Loop residues and pan-WNK Activation Loop phosphorylation sites are yellow, other colors indicate assay results, as in (A).
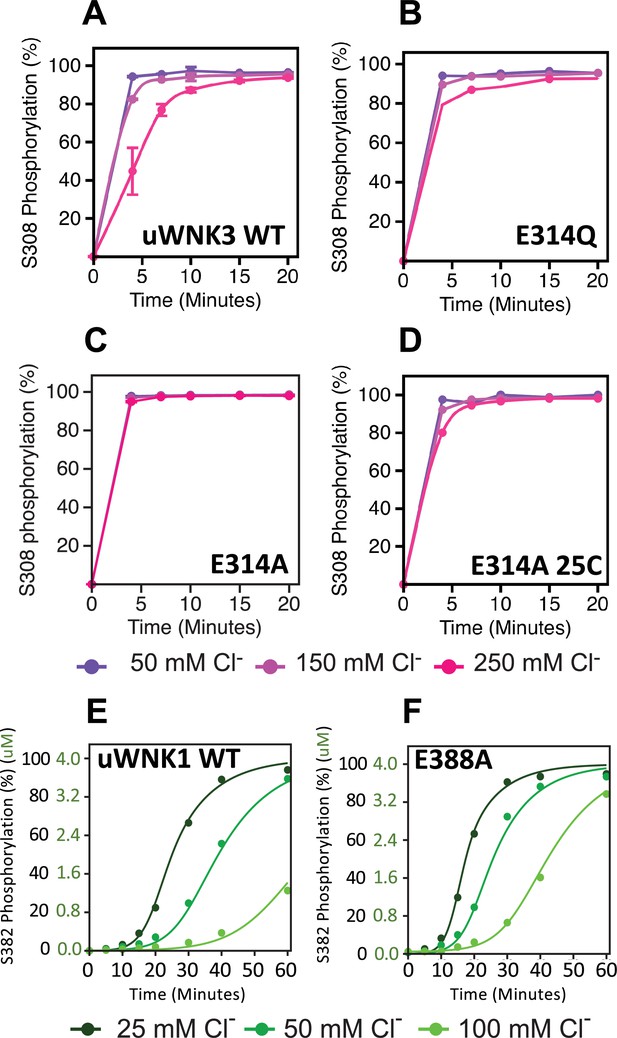
Basal autophosphorylation of WNK3/S308 and WNK1/S382 and effects of chloride on highly active mutations of position WNK3/E314 and WNK1/E388.
(A) Wild-type uWNK3. (B) uWNK3/E314Q. (C) uWNK3/E314A. (D) uWNK3/E314A, 25°C. (E) uWNK1 activity and chloride inhibition. (F) uWNK1/E388A autophosphorylation activity. Lines in panels E and F are derived from DynaFit modeling and described in Methods and Supplementary file 1f. Reactions run in 4 μM uWNK3, 30°C (unless otherwise indicated), at chloride concentrations of 50 mM (purple/dark green), 150 mM (pink/green), and 250 mM (magenta/light green). Bars indicate standard error from triplicate independent experiments.
-
Figure 5—source data 1
Mass spectrometric quantitation of activation loop peptides encompassing WNK3/S308 or WNK1/S382 over time for wild-type and AL-CL Cluster mutants as a function of [Chloride].
- https://cdn.elifesciences.org/articles/88224/elife-88224-fig5-data1-v1.xlsx
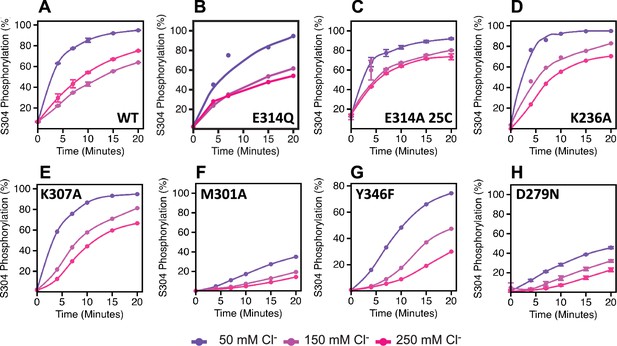
Effects of chloride on uWNK3 AL-CL Cluster mutant autophosphorylation at S304.
(A) Wild-type uWNK3. (B) uWNK3/E314Q. (C) uWNK3/E214A at 25°C. (D) uWNK3/K236A. (E) uWNK3/K307A. (F) uWNK3/M301A. (G) uWNK3/Y346F. (H) uWNK3/D279N. Reactions run in 4 μM uWNK3, 30°C (unless otherwise indicated), at sodium chloride concentrations of 50 mM (purple), 150 mM (pink), and 250 mM (magenta). Bars indicate standard error from triplicate independent experiments.
-
Figure 5—figure supplement 1—source data 1
Mass spectrometric quantitation of WNK3/S304 phosphorylation over time in chloride.
- https://cdn.elifesciences.org/articles/88224/elife-88224-fig5-figsupp1-data1-v1.xlsx
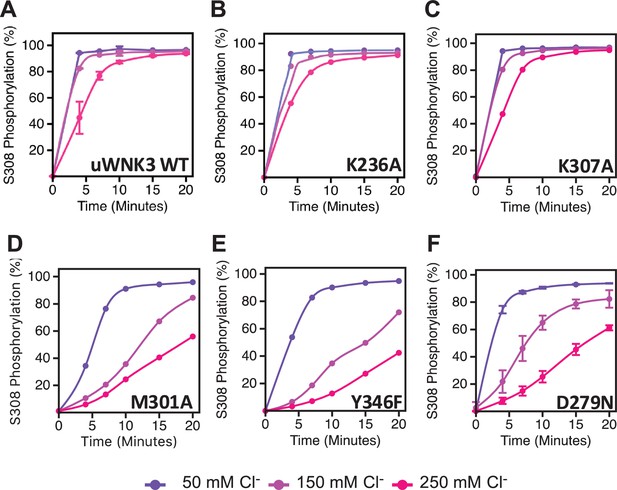
Basal autophosphorylation activity and effects of chloride on uWNK3 AL-CL Cluster mutants either similar to WT uWNK3 or less active.
(A) Wild-type uWNK3. (B) uWNK3/K236A. (C) uWNK3/K307A. (D) uWNK3/M301A. (E) uWNK3/Y346F. (F) uWNK3/D279N. Reactions run in 4 μM uWNK3, 30°C at chloride concentrations indicated. S308 phosphorylation. Bars indicate standard error from triplicate independent experiments.
-
Figure 6—source data 1
Mass spectrometric quantitation of WNK3/S308 phosphorylation for less active mutants.
- https://cdn.elifesciences.org/articles/88224/elife-88224-fig6-data1-v1.xlsx
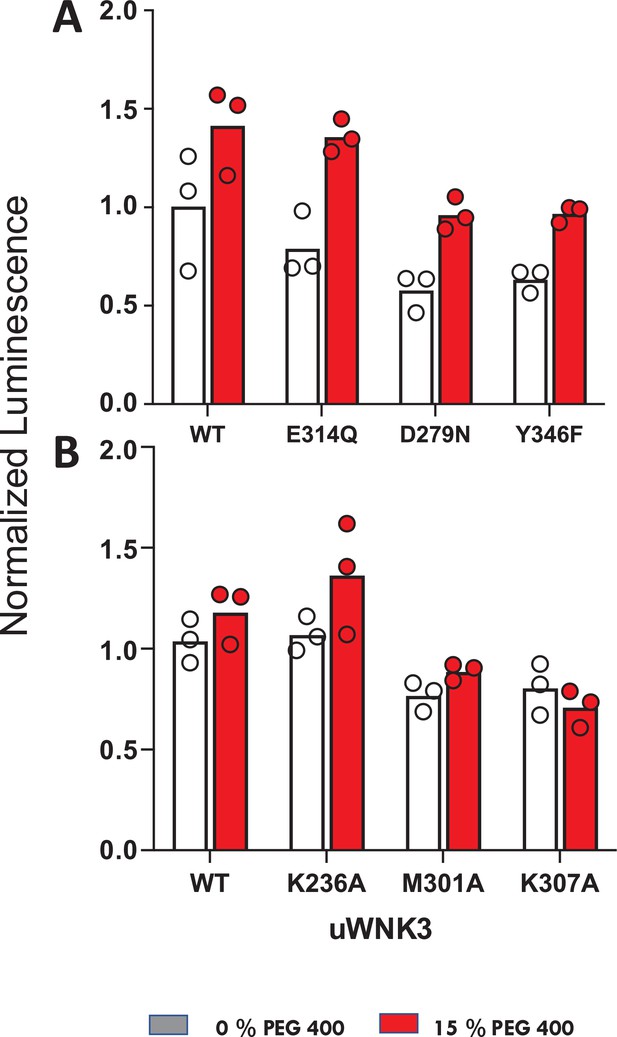
Effects of osmolytes on uWNK3 AL-CL Cluster mutant substrate phosphorylation.
(A) WT-uWNK3, E314Q, D279N, and Y346F. (B) WT-uWNK3, K236A, M301A, and K307A. ADP Glo (Promega) with gOSR1 peptide as a substrate in the absence (gray) or presence (red) of 15% PEG400 15 min, 25°C.
-
Figure 7—source data 1
Activity of WNK3 mutants toward gOSR1 as measured with ADP-Glo.
- https://cdn.elifesciences.org/articles/88224/elife-88224-fig7-data1-v1.xlsx
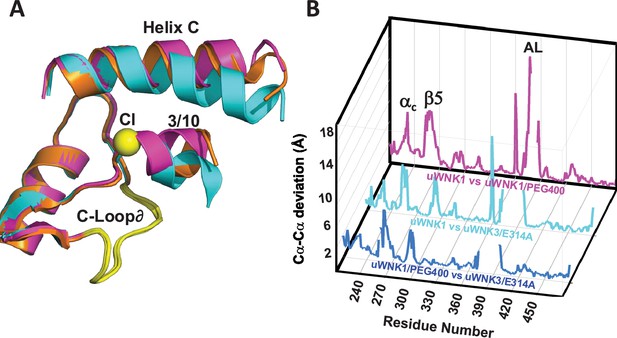
WNK3/SA/E314A structure overlay.
(A) Colors the same as Figure 3, 6CN9 (cyan), uWNK1/SA/PEG400 (PDB file 9D3F, magenta), and WNK3/E314A (PDB file 9D7Q, orange). (B) Conformational differences as a function of sequence between uWNK1 and uWNK1/SA/PEG400 (pink), uWNK1 and uWNK3/SA/E314A (cyan), and uWNK1/SA/PEG400 and WNK3/SA/E314A (blue). Changes in Activation Loop residues have been omitted because they are off-scale due to disorder in PEG400 and domain-swapping in WNK3/E314A.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Recombinant DNA reagent | WNK1(194-483) (plasmid) | This paper | GenScript | |
| Recombinant DNA reagent | WNK1(194-483)_E388A (plasmid) | This paper | GenScript | |
| Recombinant DNA reagent | WNK3(118-409) (plasmid) | Akella et al., 2021 | GenScript | |
| Recombinant DNA reagent | WNK3(118-409)_K236A (plasmid) | This paper | GenScript | |
| Recombinant DNA reagent | WNK3(118-409)_D279N (plasmid) | This paper | GenScript | |
| Recombinant DNA reagent | WNK3(118-409)_M301A (plasmid) | This paper | GenScript | |
| Recombinant DNA reagent | WNK3(118-409)_K307A (plasmid) | This paper | GenScript | |
| Recombinant DNA reagent | WNK3(118-409)_E314Q (plasmid) | This paper | GenScript | |
| Recombinant DNA reagent | WNK3(118-409)_E314A (plasmid) | This paper | GenScript | |
| Recombinant DNA reagent | WNK3(118-409)_Y346F (plasmid) | This paper | GenScript | |
| Recombinant DNA reagent | WNK3(118-409)_S308A_E314A (plasmid) | This paper | GenScript | |
| Recombinant DNA reagent | PP1cΥ (plasmid) | Barford and Keller, 1994 | From Depaoli-Roach | |
| Peptide, recombinant protein | GST-OSR1(314-344) | Taylor et al., 2018 | From Melanie Cobb | |
| Peptide, recombinant protein | Lambda phosphatase | Santa Cruz Biotechnology | Cat# sc-200312 | |
| Commercial assay or kit | ADP-Glo Max Assay | Promega | Cat# V7001 |
Additional files
-
Supplementary file 1
Tables.
(a) Crystallographic data and refinement of WNK1/S382A and WNK1/SA/PEG400. (b) Cell constant superposition comparisons. (c) WNK3/1 expression level, and Activation Loop S308/S382 phosphorylation and activity. (d) WNK1 and WNK3 peptides monitored by liquid chromatography–mass spectrometry (LC–MS). (e) uWNK1 and uWNK3 autophosphorylation assay conditions. (f) Wild-type and E388A uWNK1 kinetic model. (g) Molecular weight versus [WNK] by static light scattering. (h) Crystallographic data and refinement of WNK3/SA/E314A.
- https://cdn.elifesciences.org/articles/88224/elife-88224-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/88224/elife-88224-mdarchecklist1-v1.docx





