Understanding genetic variants in context
Figures
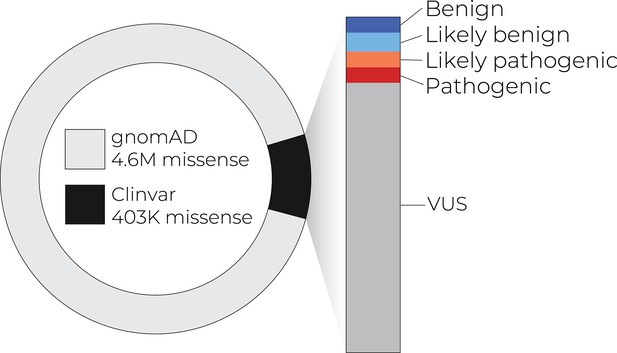
Only a small number of coding variants have annotations that can guide diagnosis and treatment.
As exome and whole-genome sequencing becomes commonplace in the clinic, the number of variants of uncertain significance is likely to increase.
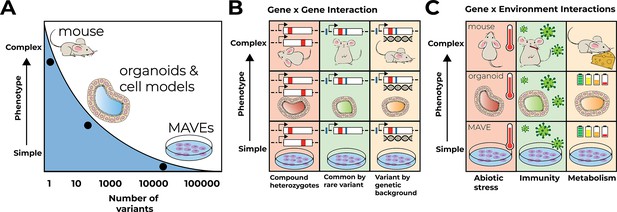
Multiplex assays of variant effect (MAVEs) in context.
(A) MAVEs in cell lines can assay many variants for simple phenotypes like cell growth. Models like organoids and mice allow for measuring complex multicellular phenotypes like proportions of cell types but are currently limited to assaying only a few variants at a time. (B) Gene–gene interactions are examined in different models at different levels of phenotype complexity. Gene–gene interactions suggested for prioritization include compound heterozygotes, combinations of common and rare variants in a given locus in cis and trans, and experiments testing variants on different genetic backgrounds. (C) Gene–environment interactions are examined at different levels of phenotype complexity. Three broad categories are suggested to model the complexity of environmental context in the laboratory: abiotic stress, challenges to immunity, and metabolism.
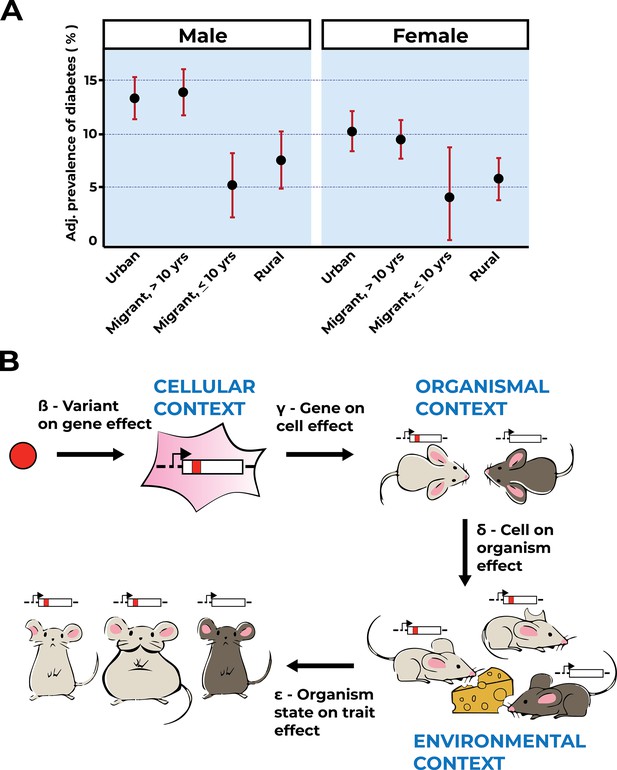
Environmental context is key to trait interpretation.
(A) Adapted from Figure 1 of Ebrahim et al., 2010, age-, factory-, and occupation-adjusted percent prevalence (95% CI) of diabetes by type of migrant and sex, Indian migration study 2005–2007. Diabetes is prevalent in urban residents and residents who migrated to urban areas and resided there for more than 10 years. (B) Gene–environment interactions will affect an organismal trait at the level of genes, cells, tissues, and whole organisms. Extending the Mostafavi et al., 2022 model to incorporate environmental context captures more relevant biology, and hence facilitates variant effect interpretation. As shown, a variant (red allele) affects a gene’s function within a particular cellular context. Cells affected by the red allele function exist within the context of the organism, here a light-gray mouse as compared to a dark-gray mouse that does not carry the red allele. Mice with or without the red allele are exposed to environmental factors, symbolized by the cheese as a dietary factor challenging metabolism. Note that one of the mice carrying the red variant is not exposed to this environmental challenge (light gray, clipped ear). In the example shown, environmental context determines the trait value (obesity, big light-gray mouse) for mice carrying the red variant.





