PITAR, a DNA damage-inducible cancer/testis long noncoding RNA, inactivates p53 by binding and stabilizing TRIM28 mRNA
Figures
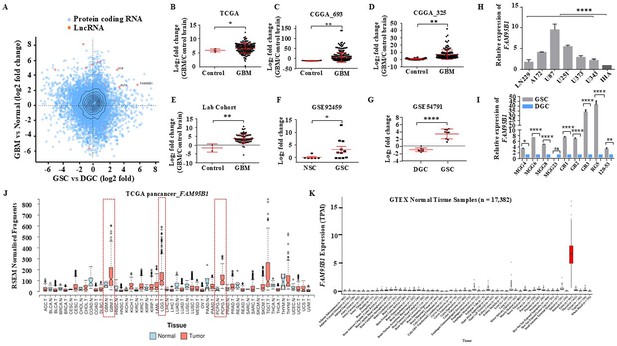
Identifying Glioblastoma stemcell-specific and conserved cancer/testis LncRNA.
(A) The Scatterplot depicts the differentially regulated protein-coding (Blue dots) and lncRNA (Red dots) transcripts. The X-axis shows a differentially regulated gene (log2 fold change) in GBM vs. normal (TCGA patient cohort), and the Y-axis represents differentially regulated genes (log2 fold change) in the GSC Vs. DGC dataset (GSE54791). (B–D) The expression in log2 fold change of FAM95B1 (PITAR) was shown in TCGA and two patient cohorts of the CGGA dataset (CGGA_693 & CGGA_325). GlioVis was used to obtain the gene expression matrix, and a t-test was performed using GraphPad Prism v6. (E) Plot depicts log2 fold change of FAM95B1 (PITAR) in our patient cohort (normal, n=3 and GBM, n=79). Data are shown as mean ± SD and an unpaired t-test was performed using GraphPad Prism v6 (**p-value <0.01). (F) Expression of PITAR in GSC vs. NSC dataset (GSE92459). (G) Expression of FAM95B1 (PITAR) in GSC vs. DGC dataset (GSE54791). Data are shown as mean ± SD and a unpaired t-test was performed using GraphPad Prism v6 (*p-value <0.05, ****p-value <0.0001). (H) Relative expression of FAM95B1 (PITAR) was quantified by qRT-PCR in different Glioblastoma cell lines and immortalized human astrocytes (IHA). Data are shown as mean ± SD (n=3) and statistically analyzed with one-way ANOVA (****p-value <0.0001). (I) Relative expression of FAM95B1 (PITAR) was measured in seven GSCs and their corresponding DGCs using the qRT-PCR method. Data are shown as mean ± SD (n=3) and an unpaired t-test was performed using GraphPad Prism v6 (*p-value <0.05, ****p-value <0.0001). (J) Relative expression (RPKM) of FAM95B1 (PITAR) across different cancer types in the TCGA Pan-cancer cohort. (K) Expression in TPM of FAM95B1 (PITAR) amongst the GTEx normal bulk tissue RNA-seq dataset. Data are shown as mean ± SD and a unpaired t-test was performed using GraphPad Prism v6 (*p-value <0.05, **p-value <0.01, ****p-value <0.0001).
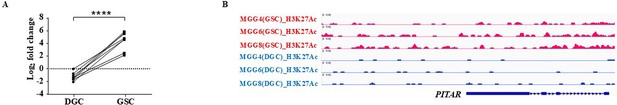
Glioblastoma stem cell-specific expression of FAM95B1 (PITAR).
(A) The plot depicts one to one correlation between GSCs and their corresponding DGCs and a paired t-test was performed using GraphPad Prism v6 (****p-value <0.0001).(B) The genomic track shows active histone marks (H3K27Ac) in GSCs over DGCs derived from the GSE54791 dataset using the IGV tools.
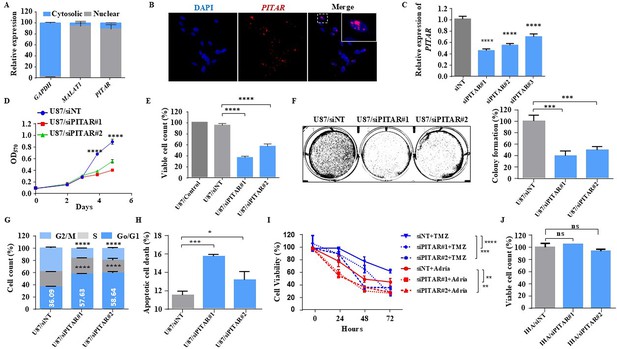
Glioblastoma cell proliferation and chemosensitivity altered by PITAR silencing.
(A) Plot depicts subcellular fractionation of U87 cells followed by quantification using qRT-PCR. MALAT1 is a positive control for nuclear gene expression, and GAPDH is a positive control for cytoplasmic gene expression. Data are shown as mean ± SD (n=3) and an unpaired t-test was performed using GraphPad Prism v6 (*p-value <0.05, **p-value <0.01, ****p-value <0.0001). (B) The RNAScope images of PITAR (red) and the DAPI (nucleus, blue) counterstain in U87 cells. The indicative scale bar on the images is 50 µm. (C) The knockdown efficiency of siRNAs (siPITAR#1, siPITAR#2, and siPITAR#3) against PITAR was measured by qRT-PCR. Data are shown as mean ± SD (n=3) and an unpaired t-test was performed using GraphPad Prism v6 (****p-value <0.0001). (D) The cell proliferation was measured by MTT assay upon PITAR knockdown in U87 cells. (E) The viable cell count was measured using a viable cell counter following the Trypan blue method. Data are shown as mean ± SD (n=3) and an unpaired t-test was performed using GraphPad Prism v6 (****p-value <0.0001). (F) Colony formation assay was performed upon PITAR knockdown compared to siNT in U87 cells. Data are shown as mean ± SD (n=3) and a unpaired t-test was performed using GraphPad Prism v6 (***p-value <0.001). (G) Cell cycle analysis was performed in PITAR-silenced U87 cells. Data are shown as mean ± SD (n=3) and an unpaired t-test was performed using GraphPad Prism v6 (****p-value <0.0001). (H) The apoptotic cell death was measured by Annexin V/PI staining in PITAR-silenced U87 cells. Data are shown as mean ± SD (n=3) and an unpaired t-test was performed using GraphPad Prism v6 (*p-value <0.05, ***p-value <0.001). (I) The chemosensitivity upon PITAR silencing was measured by MTT assay against Adriamycin (0.25 µg/ml) and Temozolomide (300 µM) in U87 cells compared to control cells. Data are shown as mean ± SD (n=3) and an unpaired t-test was performed using GraphPad Prism v6 (**p-value <0.01, ***p-value <0.001, ****p-value <0.0001). (J) The viable cell count of human astrocytes (IHA) was measured upon PITAR silencing. Data are shown as mean ± SD (n=3) and “ns” represents not significant.
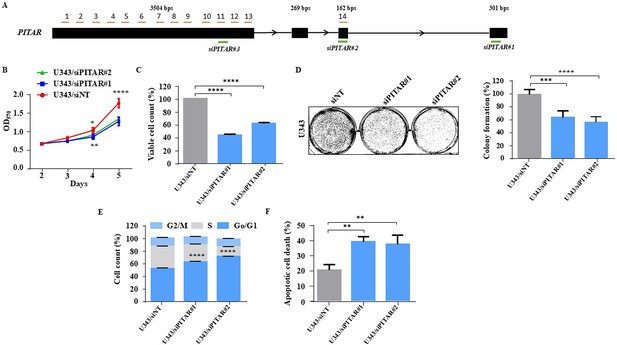
Silencing of PITAR inhibits cell growth in U343 glioma cells.
(A) Schematic of Odd probes, Even probes, and siRNAs location map with reference to the PITAR transcript. (B) Cell proliferation was measured by using an MTT assay upon PITAR knockdown compared to control U343 cells. (C) Viable cell count was measured using a viable cell counter following the Trypan blue method in U343 cells. (D) Colony formation assay was performed upon PITAR knockdown condition compared to control U343 cells. (E) Cell cycle analysis was carried out in PITAR-silenced U343 cells. (F) The apoptotic cell death was measured by Annexin V/PI staining in PITAR-silenced U343 cells. Data are shown as mean ± SD (n=3) and an unpaired t-test was performed using GraphPad Prism v6 (*p-value <0.05, **p-value <0.01, ***p-value <0.001, ****p-value <0.0001).
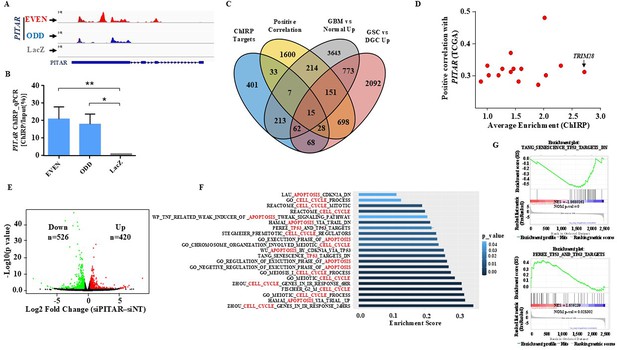
Identification of PITAR targets.
(A) Genomic track for PITAR derived from ChIRP-RNA sequencing using Odd, Even, and LacZ antisense probe. (B) PITAR Pulldown by ChIRP assay was quantified using qRT-PCR. (C) The Venn diagram represents the association of four datasets (ChIRP enriched genes, PITAR positive correlated genes from TCGA, GBM vs. normal upregulated genes from TCGA, and GSC vs. DGC upregulated genes from GSE54791). (D) The selected 15 genes from the Venn diagram are plotted in the scatter plot, and an arrow marked TRIM28 as a selected target. (E) The volcano plot depicts up-regulated (n=420) and down-regulated (n=526) genes upon PITAR knockdown compared to siNT. The gene expression matrix between siPITAR and siNT was used to construct a volcano plot to visualize differentially expressed genes. (F) Gene set enrichment analysis (GSEA) of differentially regulated genes was performed based on PITAR expression level at log2fold >0.58 and p<0.05. (G) The GSEA plots depict the enrichment of p53 up and down target gene sets, results derived from PITAR-silenced U87 cells. Data are shown as mean ± SD (n=3). ***p-value <0.001, **p-value <0.01, *p-value <0.05.
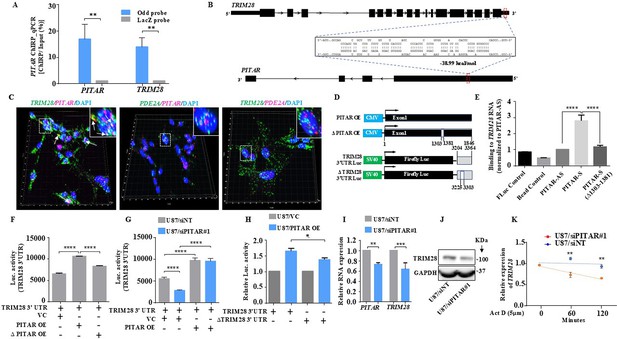
PITAR regulates the expression of TRIM28 by physical interaction with TRIM28.
(A) The qRT-PCR of PITAR and TRIM28 RNA was performed in ChIRP-RNA pull-down samples. The Probes for Odd and LacZ were used to pull down endogenous PITAR and interacting TRIM28 mRNA in U87 cells. (B) Schematic represents the predicted RNA–RNA interaction between PITAR and the 3′ UTR of TRIM28. (C) RNAScope images of co-localized signals of PITAR (red) and TRIM28 (green) in U87 cells. The panel shows the 3D reconstructed cell images of the merged 2D image (Imaris image analysis software). Yellow dots shown by white arrows depict co-localized PITAR (red) and TRIM28 (green). Magnified co-localized puncta were shown in the inset at the upper right corners, indicated by a white dotted box. The panel shows the RNAScope images of the localization of PITAR (red), TRIM28 (green), and PDE2A (green/red). PDE2A RNA was used as a negative control. The indicative scale bar on the images is 50 µm. (D) Schematic of vector plasmid construct for PITAR OE, ΔPITAR OE, TRIM28 3′ UTR, and ΔTRIM28 3′ UTR. (E) PITAR interaction with the TRIM28 3′ UTR was measured using an in vitro RNA–RNA interaction assay and compared to a panel of control RNAs (PITAR antisense, Fluc control, Bead control). The binding affinity was quantified by qRT-PCR analysis of the TRIM28. Data were normalized to the PITAR-AS control. (F) The Luc activity of TRIM28 3′ UTR was measured after the ectopic expression of PITAR and ΔPITAR in U87 cells using a luciferase reporter assay. (G) Luciferase assay was performed in PITAR silenced U87 cells co-transfected with VC and PITAR OE vector. (H) The Firefly luciferase activity was measured in U87 cells containing a deleted PITAR binding site of TRIM28 3’UTR (ΔTRIM28 3′ UTR), co-transfected with VC and PITAR OE. (I) Relative expression of TRIM28 in PITAR-silenced U87 cells was measured by qRT-PCR. (J) The TRIM28 protein expression was measured by immunoblotting. Data are shown as mean ± SD (n=3) and an unpaired t-test was performed using GraphPad Prism v6 (*p-value <0.05, **p-value <0.01, ****p-value <0.0001). (K) TRIM28 transcript was measured at indicated time points post Actinomycin D (5 μg/ml) treatment in siNT and siPITAR-transfected U87 cells by qRT-PCR (n=3). The log2 ratio of the remaining TRIM28 was plotted using linear regression after normalizing to the 0th hour of the respective condition using GraphPad Prism v6. Data are shown as mean ± SD (**p-value <0.01).
-
Figure 4—source data 1
Raw unedited blots for Figure 4J.
- https://cdn.elifesciences.org/articles/88256/elife-88256-fig4-data1-v1.zip
-
Figure 4—source data 2
Uncropped and labeled blots for Figure 4J.
- https://cdn.elifesciences.org/articles/88256/elife-88256-fig4-data2-v1.pdf
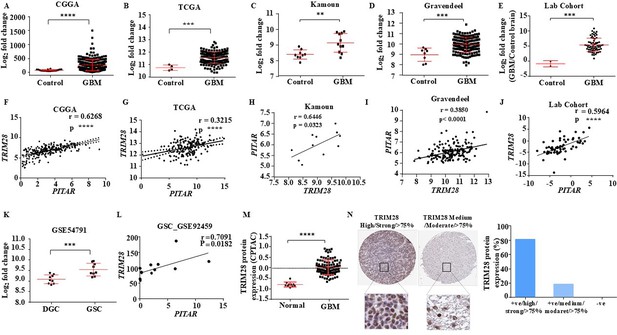
Clinical relevance of PITAR and TRIM28 association.
(A–D) Expression in log2 fold change of TRIM28 in Normal vs. GBM, derived from the CGGA, TCGA, Kamoun, and Gravendeel dataset. GlioVis was used to obtain the gene expression matrix, and an unpaired t-test was performed using GraphPad Prism v6 (**p-value <0.01, ***p-value <0.001, ****p-value <0.0001). (E) Expression in log2 fold change of TRIM28 in our lab patient cohorts is quantified using qRT-PCR. Data are shown as mean ± SD and an unpaired t-test was performed using GraphPad Prism v6 (***p-value <0.001). (F–J) The correlation analysis was performed between PITAR and TRIM28 in glioma samples from the CGGA, TCGA, Kamoun, and Gravendeel databases and our lab patient cohort. The correlation coefficient was calculated using Pearson’s correlation coefficient test using GraphPad Prism v6 (**p-value <0.01, ***p-value <0.001, ****p-value <0.0001). (K) Expression in log2 fold change of TRIM28 was quantified in GSC and corresponding DGC from the GSE54791 database. Data are shown as mean ± SD and an unpaired t-test was performed using GraphPad Prism v6 (***p-value <0.001). (L) The correlation analysis between PITAR and TRIM28 in GSC is derived from the GSE92459 database. The correlation coefficient was calculated using Pearson’s correlation coefficient test using GraphPad Prism v6 (*p-value <0.05). (M) TRIM28 protein expression in Normal vs. GBM, derived from the CPTAC database. (N) TRIM28 protein expression was quantified using immunohistochemistry images from the Protein atlas. Data are shown as mean ± SD and an unpaired t-test was performed using GraphPad Prism v6 (****p-value <0.0001).
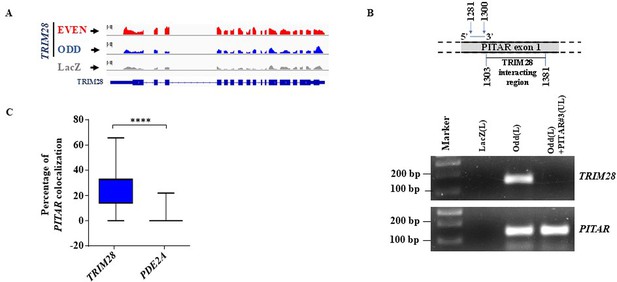
PITAR physically interacts with TRIM28 mRNA.
(A) Genomic track for TRIM28 derived from ChIRP-RNA sequencing using Odd, Even, and LacZ antisense probes. (B) A competition assay based on antisense oligonucleotide blocking was performed upon fragmented total RNA, preincubated with unlabeled PITAR antisense probe #3 (PITAR#3(UL)), which is located close to the TRIM28 binding region on the exon 1 of PITAR, followed by pulldown with the PITAR biotinylated antisense (odd(L)) probe set of PITAR and LacZ biotinylated antisense (LacZ(L)) was employed as a negative control for the pulldown experiment. (C) RNAScope images of co-localized signals were quantified using Imaris microscopy image analysis software 9.8.2 version. Data are shown as mean ± SD and an unpaired t-test was performed using GraphPad Prism v6 (****p-value <0.0001).
-
Figure 4—figure supplement 2—source data 1
Raw unedited gels for Figure 4—figure supplement 2B.
- https://cdn.elifesciences.org/articles/88256/elife-88256-fig4-figsupp2-data1-v1.zip
-
Figure 4—figure supplement 2—source data 2
Uncropped and labeled gels for Figure 4-figure supplement 2B.
- https://cdn.elifesciences.org/articles/88256/elife-88256-fig4-figsupp2-data2-v1.pdf
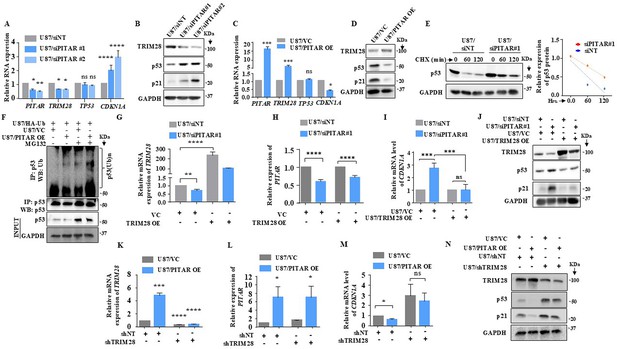
PITAR regulates wild-type p53 protein levels via TRIM28-mediated ubiquitination.
(A) Relative expression of PITAR, TRIM28, TP53, and CDKN1A was quantified by qRT-PCR in PITAR-silenced U87 cells compared to siNT. (B) The protein expression of TRIM28, p53, and p21 was measured by immunoblotting in PITAR-silenced U87 cells compared to siNT. (C, D) Cells were transfected with pcDNA3.1-PITAR (PITAR OE)/ empty vector control plasmid (pcDNA3.1) and harvested 48 hr post-transfection for qRT-PCR (PITAR, TRIM28, TP53, and CDKN1A) and immunoblotting with indicated antibodies (TRIM28, p53, and p21). GAPDH served as the control. (E) The Half-life of the p53 protein was measured in PITAR-silencing (siPITAR) and control (siNT) U87 cells with the treatment of cycloheximide (CHX; 50 μg/mL). The relative expression of the remaining p53 was plotted using linear regression after normalizing to the 0th hour of the respective condition. (F) The endogenous level of p53 ubiquitination was measured in pcDNA3.1-PITAR (PITAR OE)/empty vector plasmid (pcDNA3.1) stable U87 cells by p53 immunoprecipitation followed by immunoblotting with the indicated antibodies in the presence and absence of MG132. (G–I) The relative expression of PITAR, TRIM28, and CDKN1A was measured by qRT-PCR in U87/siPITAR#1 and U87/siNT cells with exogenously overexpressed TRIM28 conditions. (J) The protein expression of TRIM28, p53, and p21 was measured by immunoblotting in U87/siPITAR#1 and U87/siNT cells with exogenously overexpressed TRIM28 condition. (K–M) The relative expression of PITAR, TRIM28, and CDKN1A was measured by qRT-PCR in U87/shTRIM28 and U87/shNT cells with exogenously overexpressed PITAR conditions. (N) The protein expression of TRIM28, p53, and p21 was measured by immunoblotting in U87/shTRIM28 and U87/shNT cells with exogenously overexpressed PITAR conditions. Data are shown as mean ± SD (n=3) and an unpaired t-test was performed using GraphPad Prism v6 (*p-value <0.05, **p-value <0.01,***p-value <0.001, ****p-value <0.0001).
-
Figure 5—source data 1
Raw unedited blots for Figure 5.
- https://cdn.elifesciences.org/articles/88256/elife-88256-fig5-data1-v1.zip
-
Figure 5—source data 2
Uncropped and labeled blots for Figure 5.
- https://cdn.elifesciences.org/articles/88256/elife-88256-fig5-data2-v1.pdf
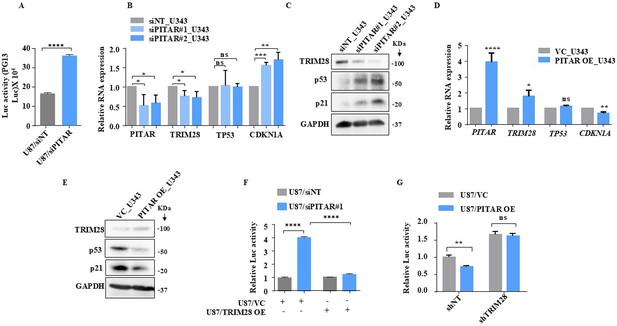
PITAR inhibits wild-type p53 protein through TRIM28.
(A) The PG13-Luc activity was measured in PITAR-silenced/siNT U87 cells. (B) Relative expression of PITAR, TRIM28, TP53, and CDKN1A was quantified by qRT-PCR in PITAR-silenced U343 cells compared to siNT. (C) The protein expression of TRIM28, p53, and p21 was measured by immunoblotting in PITAR-silenced U343 cells compared to siNT. (D, E) Cells were transfected with pcDNA3.1-PITAR (PITAR OE)/ empty vector control plasmid (pcDNA3.1) and harvested 48 hr post-transfection for qRT-PCR (PITAR, TRIM28, TP53, and CDKN1A) and immunoblotting with indicated antibodies (TRIM28, p53, and p21). GAPDH served as the control. (F) The PG13-Luc activity was measured in U87/siPITAR and U87/siNT cells with exogenously overexpressed TRIM28 conditions. (G) The PG13-Luc activity was measured in U87/shTRIM28 and U87/shNT cells with exogenously overexpressed PITAR conditions. Data are shown as mean ± SD (n=3) and an unpaired t-test was performed using GraphPad Prism v6 (*p-value <0.05, **p-value <0.01,***p-value <0.001, ****p-value <0.0001).
-
Figure 5—figure supplement 1—source data 1
Raw unedited blots for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/88256/elife-88256-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
Uncropped and labeled blots for Figure 5-figure supplement 1.
- https://cdn.elifesciences.org/articles/88256/elife-88256-fig5-figsupp1-data2-v1.pdf
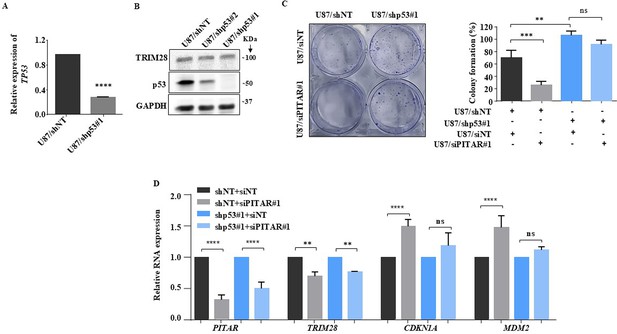
Inhibition of glioma cell growth by PITAR requires wild type p53.
(A) The relative expression of p53 was quantified in U87/shNT and U87/shp53 cells by qRT-PCR. (B) The immunoblot represents the knockdown efficiency of two shp53 clones and the expression of TRIM28 in the p53 silenced condition. (B) The colony formation assay was performed and quantified in siNT and siPITAR conditions in a p53 knockdown background. (D) The relative expression of PITAR, TRIM28, CDKN1A, and MDM2 was quantified in siNT and siPITAR conditions in a p53 Knockdown background using qRT-PCR. Data are shown as mean ± SD (n=3) and an unpaired t-test was performed using GraphPad Prism v6 (*p-value <0.05, **p-value <0.01,***p-value <0.001, ****p-value <0.0001).
-
Figure 5—figure supplement 2—source data 1
Raw unedited blots for Figure 5—figure supplement 2B.
- https://cdn.elifesciences.org/articles/88256/elife-88256-fig5-figsupp2-data1-v1.zip
-
Figure 5—figure supplement 2—source data 2
Uncropped and labeled blots for Figure 5-figure supplement 2B.
- https://cdn.elifesciences.org/articles/88256/elife-88256-fig5-figsupp2-data2-v1.pdf
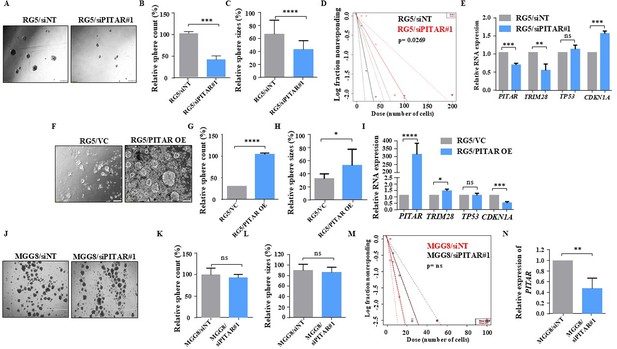
Glioblastoma stem-like cell growth is induced by PITAR through p53 inactivation.
(A) The bright field image represents RG5 sphere growth in the siNT/siPITAR#1 condition. (B, C) RG5 sphere growth was quantified by the measurement of sphere count and sphere size using ImageJ (https://imagej.nih.gov/ij/download.html). (D) Limiting dilution assay was performed in RG5 cells. The graph represents the percentage of wells without spheres as a function of the number of PITAR Knockdown cells compared to control cells (https://bioinf.wehi.edu.au/software/elda/). (E) Relative expression of PITAR, TRIM28, TP53, and CDKN1A was quantified by qRT-PCR in PITAR-silenced RG5 cells compared to siNT. (F) The bright field image represents RG5 sphere growth in PITAR OE compared to the vector control. (G, H) RG5 sphere growth was quantified by measuring sphere count and size in the PITAR OE condition compared to the vector control using ImageJ software. (I) Relative expression of PITAR, TRIM28, TP53, and CDKN1A was quantified by qRT-PCR in the PITAR OE condition of RG5 cells compared to the vector control. (J) The bright field image represents MGG8 sphere growth in the siNT/siPITAR condition. (K, L) MGG8 sphere growth was quantified by the measurement of sphere count and sphere size using ImageJ tools. (M) Limiting dilution assay was performed in MGG8 cells. The graph represents the percentage of wells without spheres as a function of the number of PITAR Knockdown cells compared to control cells. (N) The silencing efficiency of PITAR in MGG8 cells was measured using qRT-PCR. Data are shown as mean ± SD (n=3) and an unpaired t-test was performed using GraphPad Prism v6 (*p-value <0.05, **p-value <0.01,***p-value <0.001, ****p-value <0.0001).
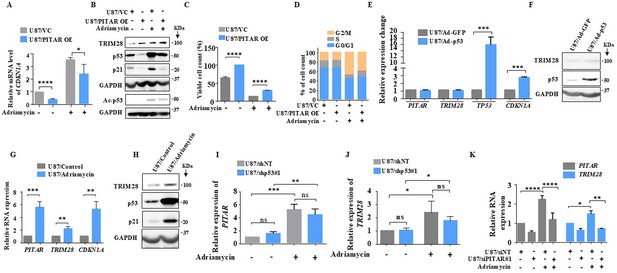
DNA damage-induced PITAR diminishes the DNA damage response by p53 through TRIM28.
(A) The relative expression of CDKN1A was measured in the presence and absence of Adriamycin by qRT-PCR in PITAR OE/VC U87 cells. (B) The protein expression of TRIM28, p53, ac-p53, and p21 was measured in the presence and absence of Adriamycin by immunoblotting in PITAR OE/VC U87 cells. GAPDH served as the control. (C) The viable cell count was performed in the presence and absence of Adriamycin in PITAR OE/VC U87 cells. (D) The cell cycle analysis was performed in the presence and absence of Adriamycin in PITAR OE/VC U87 cells. (E) The relative expression of PITAR, TRIM28, TP53, and CDKN1A was measured by qRT-PCR in Ad-p53 and Ad-GFP-infected U87 cells. (F) The protein expression of TRIM28 and p53 was measured by immunoblotting in Ad-p53 and Ad-GFP-infected U87 cells. (G) The relative expression of PITAR, TRIM28, and CDKN1A was measured in the presence and absence of Adriamycin by qRT-PCR in U87 cells. (H) The immunoblot depicting the expression of TRIM28, p53, and p21 upon treatment of Adriamycin. (I, J) The qRT-PCR was performed to measure the relative expression of PITAR and TRIM28 in the p53 knockdown condition upon Adriamycin treatment. (K) The relative expression of PITAR and TRIM28 was measured by qRT-PCR in Adriamycin-treated PITAR-silenced U87 cells. Data are shown as mean ± SD (n=3) and an unpaired t-test was performed using GraphPad Prism v6 (*p-value <0.05, **p-value <0.01,***p-value <0.001, ****p-value <0.0001).
-
Figure 6—source data 1
Raw unedited blots for Figure 6.
- https://cdn.elifesciences.org/articles/88256/elife-88256-fig6-data1-v1.zip
-
Figure 6—source data 2
Uncropped and labeled blots for Figure 6.
- https://cdn.elifesciences.org/articles/88256/elife-88256-fig6-data2-v1.pdf
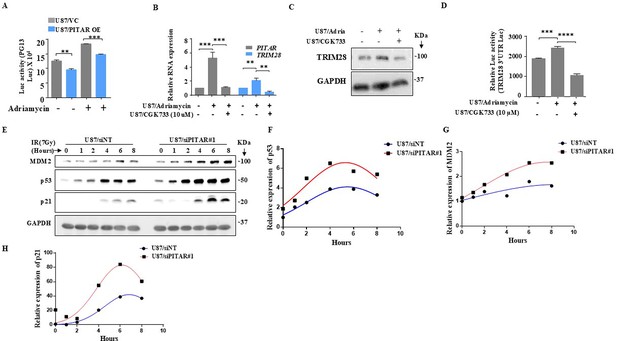
DNA damage-induced PITAR suppresses p53 respons to DNA damage.
(A) The PG13-Luc activity was measured in the presence and absence of Adriamycin in U87/PITAR OE and U87/VC cells. (B) The relative expression of PITAR and TRIM28 was measured in the presence and absence of Adriamycin and CGK733 using qRT-PCR in U87 cells. (C) The TRIM28 protein expression was measured by immunoblotting in the presence of Adriamycin and CGK733 in U87 cells. (D) The TRIM28 3’UTR Luc activity was measured in the presence of Adriamycin and CGK733 in U87 cells. Data are shown as mean ± SD (n=3) and an unpaired t-test was performed using GraphPad Prism v6 (*p-value <0.05, **p-value <0.01,***p-value <0.001, ****p-value <0.0001). (E) The U87/siNT and U87/siPITAR cells were γ-irradiated (7 Gy), and cell extracts were prepared at different time points post-irradiation as indicated. The immunoblotting for Mdm2, p53, and p21 were performed to show their kinetics. (F–H). The quantification for Mdm2, p53, and p21 immunoblot was performed and plotted with nonlinear regression curve fit.
-
Figure 6—figure supplement 1—source data 1
Raw unedited blots for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/88256/elife-88256-fig6-figsupp1-data1-v1.zip
-
Figure 6—figure supplement 1—source data 2
Uncropped and labeled blots for Figure 6-figure supplement 1.
- https://cdn.elifesciences.org/articles/88256/elife-88256-fig6-figsupp1-data2-v1.pdf
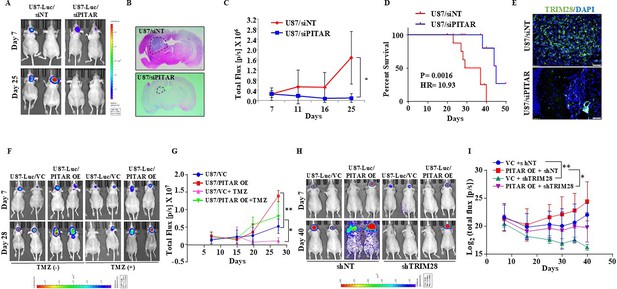
PITAR promotes glioma tumor growth and resistance to Temozolomide chemotherapy.
(A) Mice (NIH nu/nu) were injected with siNT/siPITAR#1 transfected U87-Luc cells (0.3x106 cells/mice), and tumors were allowed to grow for 50 days (n=10), and the luminescence imaging was performed using the IVIS instrument (Perkin Elmer IVIS system). (B) H&E staining was performed in formalin-fixed tumor-bearing (siNT and siPITAR#1) mouse brain sections. (C) The tumor growth curve of siNT and siPITAR#1 tumor-bearing mice was quantified over time using IVIS. The difference between groups was statistically analyzed by ANOVA with Tukey’s multiple comparisons test (*p<0.05).(D) The Kaplan–Meier graph shows the survival of mice-bearing tumors formed by siNT and siPITAR#1 cells (N=10/group), statistical differences were calculated with the Log-rank (Mantel-Cox) test (*p<0.05). (E) The mmunohistochemistry assay was performed to show the TRIM28 protein expression in the tumor tissue section derived from U87-Luc/siNT and U87-Luc/siPITAR#1 tumors. The Green color represents the TRIM28 protein, and the blue depicts the nucleus stained with DAPI. Scale bar = 100 μm. (F) Mice (NIH nu/nu) were injected with U87-Luc/PITAR OE and U87-Luc/VC cells (0.3x106 cells/mice, n=10), and tumors were allowed to grow for 30 days. The tumor-bearing mice were treated with 100 mg/kg TMZ in 25% DMSO saline solution after 11 days by intraperitoneal injection for one week, and the luminescence imaging was performed using the IVIS instrument. (G) The tumor growth curve of VC, PITAR OE, VC +TMZ, and PITAR OE +TMZ tumor-bearing mice was quantified over time using IVIS. The difference between groups was statistically analyzed by ANOVA with Tukey’s multiple comparisons test (*p<0.05, ***p<0.001). (H) Mice (NIH nu/nu) were injected with U87-Luc/PITAR OE +shNT, U87-Luc/VC +shNT, U87-Luc/PITAR OE +shTRIM28 and U87-Luc/VC +shTRIM28 cells (0.3x106 cells/mice), and tumors were allowed to grow for 50 days. (I) The tumor growth curve of VC +shNT, PITAR OE +shNT, VC +shTRIM28, and PITAR OE +shTRIM28 tumor-bearing mice (n=10) was quantified over time using IVIS. Luminescence was evaluated twice per 10 days and before sacrifice. Bars indicate standard error and the difference between groups was statistically analyzed by ANOVA with Tukey’s multiple comparisons post-test (*p<0.05, **p<0.01).
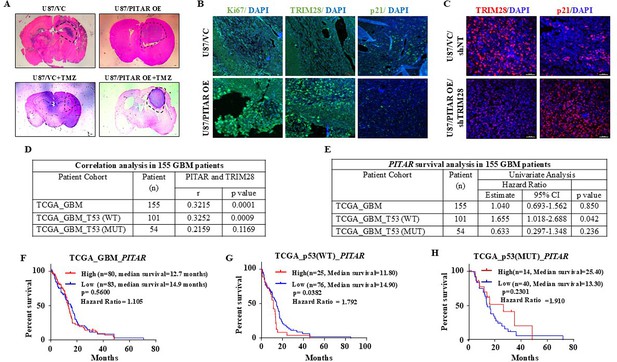
PITAR promotes tumor growth via p53 in a TRIM28-dependent manner.
(A) H&E staining was performed on formalin-fixed tumor-bearing (VC, PITAR OE, VC +TMZ, and PITAR OE +TMZ) mouse brain sections. (B) Immunohistochemistry was performed to detect TRIM28, Ki67, and p21 protein expression in tumor sections derived from VC, PITAR OE, VC +TMZ, and PITAR OE +TMZ tumors. The green color represents specific protein staining, and the blue depicts the nucleus stained with DAPI. Magnification = 20X, scale bar = 50 μm. (C) Immunohistochemistry was performed to detect TRIM28 and p21 protein expression in tumor sections derived from PITAR OE/shNT and PITAR OE/shTRIM28 tumors. The red color represents specific protein staining, and the blue depicts the nucleus stained with DAPI. Magnification = 40X, scale bar = 40 μm. (D) Correlation analysis was performed in the p53 wild type and p53 mutant GBM patient cohort. The correlation coefficient was calculated using Pearson’s correlation coefficient test using GraphPad Prism v6. (E) Univariate Cox regression analysis was performed in the p53 wild type and mutant p53 GBM patient cohort. (F–H) The Kaplan–Meier graph depicts the survival correlation of PITAR in the total GBM cohort, p53 wild type, and p53 mutant GBM patient cohort. The statistical differences were calculated with the Log-rank (Mantel-Cox) test (*p<0.05).
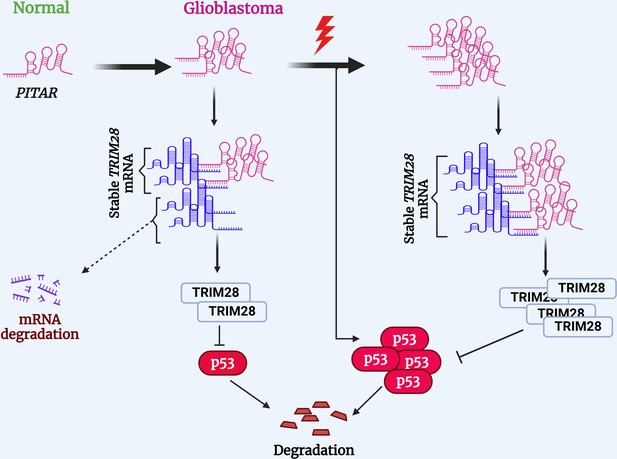
Proposed working model of this study.
PITAR inhibits p53 by binding and stabilizing TRIM28 mRNA.
© 2024, BioRender Inc. Figure 8 was created using BioRender, and is published under a CC BY-NC-ND 4.0. Further reproductions must adhere to the terms of this license
Additional files
-
Supplementary file 1
Differentially expressed Genes in GSC vs. DGC derived from GSE54791 and GBM vs. Normal derived from TCGA.
- https://cdn.elifesciences.org/articles/88256/elife-88256-supp1-v1.xlsx
-
Supplementary file 2
ChIRP enriched genes in Even, Odd and LacZ pulldown.
- https://cdn.elifesciences.org/articles/88256/elife-88256-supp2-v1.xlsx
-
Supplementary file 3
Differentially regulated genes (GBM VS Control brain) that correlate with PITAR expression.
- https://cdn.elifesciences.org/articles/88256/elife-88256-supp3-v1.xlsx
-
Supplementary file 4
Upregulated Genes in GBM – Normal from TCGA dataset.
- https://cdn.elifesciences.org/articles/88256/elife-88256-supp4-v1.xlsx
-
Supplementary file 5
Upregulated Genes in GSC – DGC from GSE54791 dataset.
- https://cdn.elifesciences.org/articles/88256/elife-88256-supp5-v1.xlsx
-
Supplementary file 6
Differentially expressed Genes in siPITAR/U87 - siNT/U87.
- https://cdn.elifesciences.org/articles/88256/elife-88256-supp6-v1.xlsx
-
Supplementary file 7
GSEA analysis of downregulated genes in siPITAR/U87 - siNT/U87.
- https://cdn.elifesciences.org/articles/88256/elife-88256-supp7-v1.xlsx
-
Supplementary file 8
In-silico prediction of RNA-RNA interaction site between TRIM28 and PITAR using IntaRNA tools.
- https://cdn.elifesciences.org/articles/88256/elife-88256-supp8-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/88256/elife-88256-mdarchecklist1-v1.docx





