YAP/TAZ enhances P-body formation to promote tumorigenesis
Figures
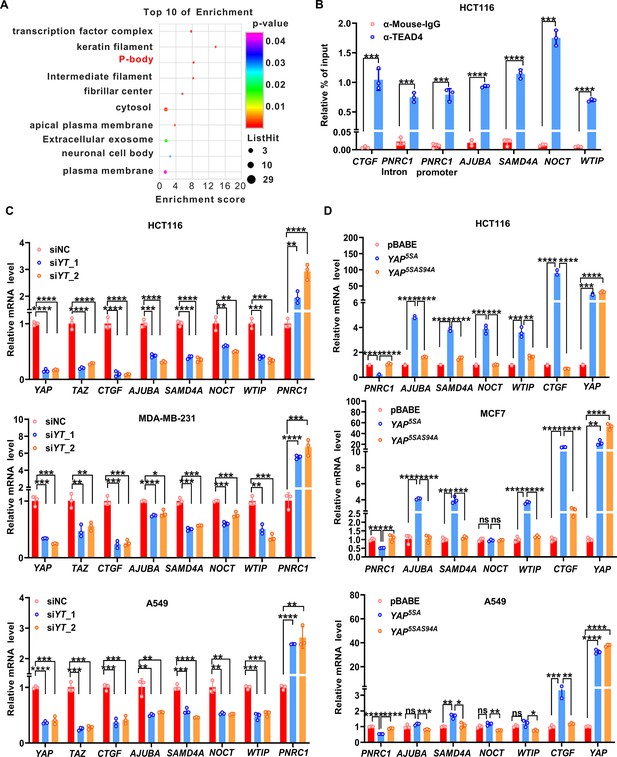
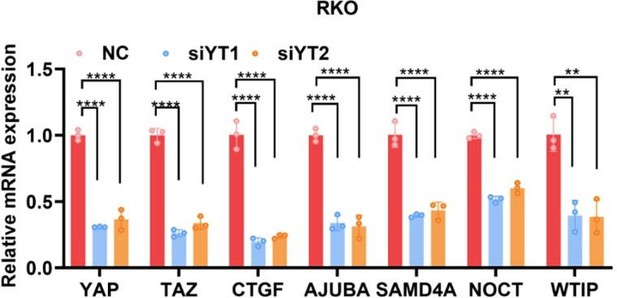
YAP/TAZ transcriptionally regulates genes related to P-bodies.
(A) Gene Ontology (GO) analysis of the downregulated genes upon knockdown of YAP/TAZ in HCT116 cells. The graph shows enrichment in the cellular component category. (B) ChIP–qPCR analysis of endogenous TEAD4 binding to the genomic locus of the indicated P-body-related genes in HCT116 cells. The CTGF promoter was included as the positive control. (C) qPCR analysis of the mRNA levels of the indicated P-body-related genes in YAP/TAZ knockdown HCT116, MDA-MB-231, and A549 cells. Cells were transfected with YAP/TAZ siRNA for 3 d before qPCR analysis. (D) qPCR analysis of the mRNA levels of the indicated P-body-related genes in HCT116, MCF7, and A549 cells stably expressing YAP5SA and YAP5SA-S94A. Cells were infected with YAP5SA- and YAP5SA-S94A-containing retroviruses and selected with puromycin for 1 wk before qPCR analysis. n = 3 biologically independent samples per group. Two-tailed Student’s t-test (B) and one-way ANOVA (C, D) were performed to assess statistical significance in this figure. These data (B–D) are representative of three independent experiments.
-
Figure 1—source data 1
Original data for the statistical analysis in Figure 1B–D.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig1-data1-v1.xlsx
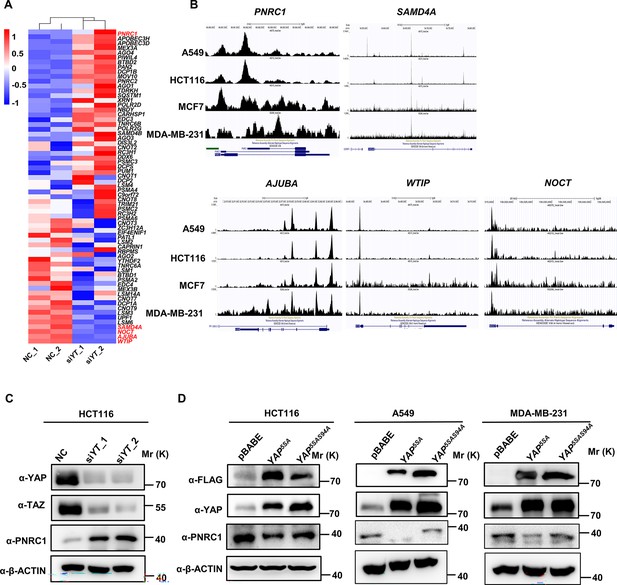
RNA-seq and ChIP-seq analysis of YAP/TEAD’s target genes related to P-bodies.
(A) Heatmap showing the mRNA levels of 76 genes annotated as P-body-related genes, as detected in HCT116 cells by RNA-seq (GSE176475). (B) Representative sequencing TEAD4 ChIP-seq tracks at the SAMD4A/AJUBA/WTIP/NCOT/PNRC1 loci in HCT-116, A549, MDA-MB-231, and MCF7 cells. The ChIP-seq data were extracted from the Cistrome database and uploaded to the UCSC Genome Browser for visualization. (C) Western blot analysis of PNRC1, YAP, and TAZ in control and YAP/TAZ knockdown HCT116 cells. (D) Western blot analysis of PNRC1, YAP, and FLAG in control HCT116 cells and HCT116 cells overexpressing FLAG-YAP5SA and YAP5SA-S94A. These data (C, D) are representative of two independent experiments.
-
Figure 1—figure supplement 1—source data 1
Original data for the heat map in Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig1-figsupp1-data1-v1.xlsx
-
Figure 1—figure supplement 1—source data 2
Original file for the Western blot analysis in Figure 1—figure supplement 1C and D.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig1-figsupp1-data2-v1.pdf
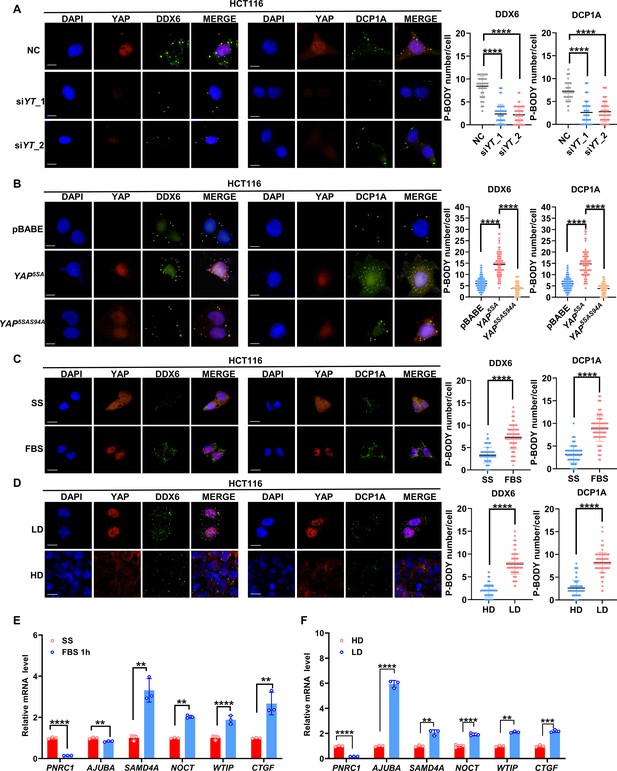
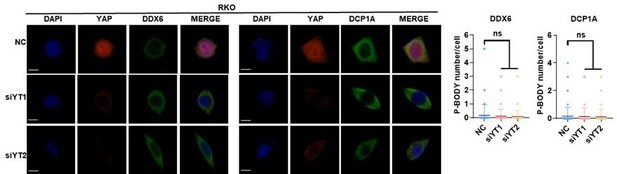
YAP/TAZ promotes P-body formation in colorectal cancer (CRC) cells.
(A) Immunofluorescence analysis of the P-body markers DDX6 and DCP1A in YAP/TAZ knockdown HCT116 cells. Cells were transfected with YAP/TAZ siRNA for 3 d before processing for immunofluorescence staining using anti-DDX6 and anti-DCP1A antibodies. Foci were counted in 100 cells per group. (B) Immunofluorescence analysis of DDX6 and DCP1A in HCT116 cells expressing YAP5SA and YAP5SA-S94A. (C) Immunofluorescence analysis of DDX6 and DCP1A in HCT116 cells. Cells were treated with 10% fetal bovine serum (FBS) for 1 hr after overnight serum starvation (SS). (D) Immunofluorescence analysis of DDX6 and DCP1A in HCT116 cells in sparse or confluent culture. (E, F) qPCR analysis of the indicated genes in HCT116 cells. HCT116 cells were treated with 10% FBS for 1 hr after overnight SD (E) or culture under sparse or confluent conditions in standard culture medium (F). Kruskal–Wallis test (A, B), Mann–Whitney U test (C, D), and two-tailed Student’s t-test (E, F) were performed to assess statistical significance. These data (A–F) are representative of three independent experiments.
-
Figure 2—source data 1
Original data for the statistical analysis in Figure 2A–F.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig2-data1-v1.xlsx
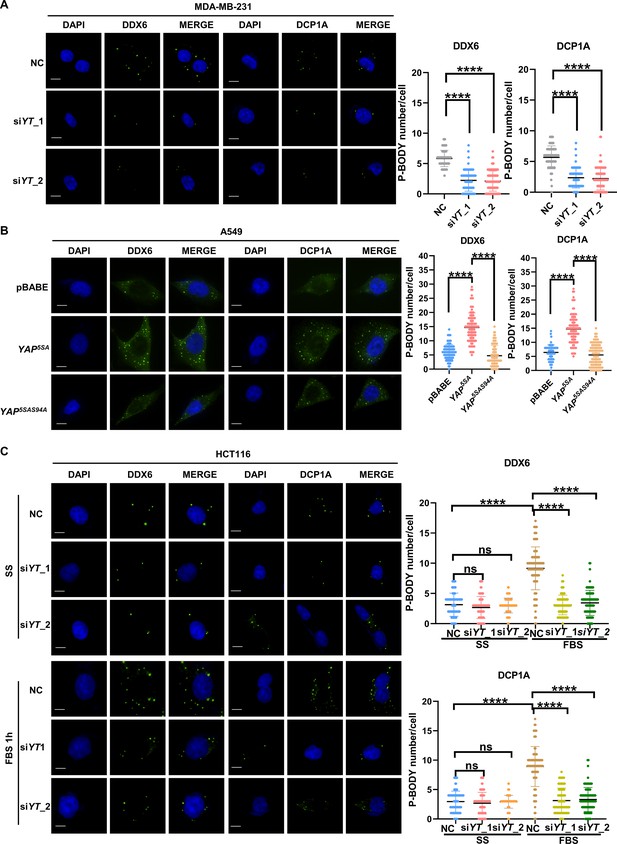
YAP/TAZ modulates P-body formation in breast, lung and colorectal cancer cells.
(A) Immunofluorescence analysis of the P-body markers DDX6 and DCP1A in YAP/TAZ knockdown MDA-MB-231 cells. Cells were transfected with YAP/TAZ siRNA for 3 d before processing for immunofluorescence staining using anti-DDX6 and anti-DCP1A antibodies. Foci were counted in 100 cells per group. (B) Immunofluorescence analysis of DDX6 and DCP1A in A549 cells expressing YAP5SA and YAP5SA-S94A. (C) Immunofluorescence analysis of DDX6 and DCP1A in HCT116 cells. Cells were transfected with control and YAP/TAZ siRNA for 3 d before overnight serum starvation. Then, the starved cells were treated with 10% fetal bovine serum (FBS) for 1 hr before immunofluorescence staining. Kruskal–Wallis test was performed to assess statistical significance. These data (A–C) are representative of three independent experiments.
-
Figure 2—figure supplement 1—source data 1
Original data for the statistical analysis in Figure 2—figure supplement 1A–C.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig2-figsupp1-data1-v1.xlsx
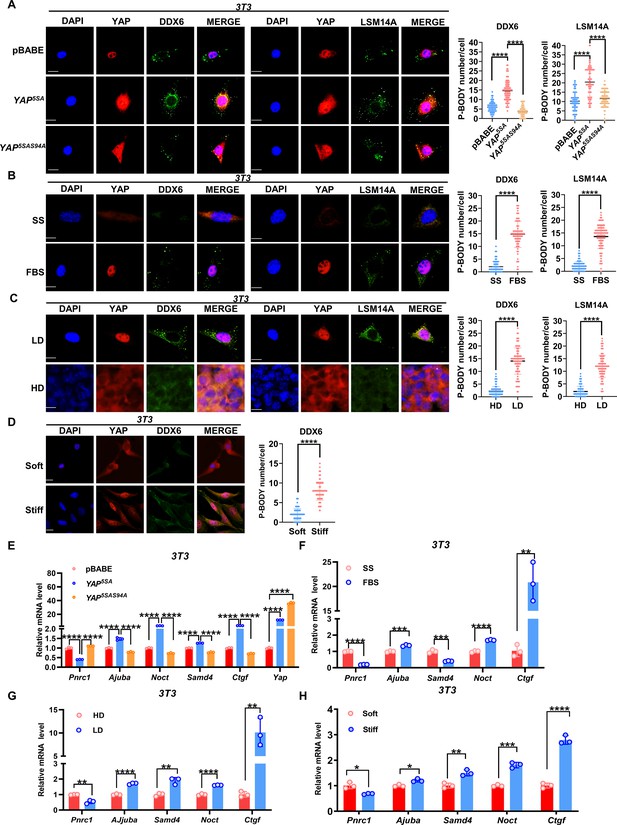
YAP/TAZ modulates P-body formation in untransformed NIH3T3 cells.
(A) Immunofluorescence analysis of DDX6 and LSM14A in NIH3T3 cells expressing YAP5SA and YAP5SA-S94A. Foci were counted in 100 cells per group. (B) Immunofluorescence analysis of DDX6 and LSM14A in NIH3T3 cells. Sparse cells were treated with 10% fetal bovine serum (FBS) for 1 hr after overnight serum starvation (SS). (C) Immunofluorescence analysis of DDX6 and LSM14A in NIH3T3 cells in sparse or confluent culture. (D) Immunofluorescence analysis of DDX6 and LSM14A in NIH3T3 cells cultured on soft (1 kPa) to stiff (40 kPa) matrices. (E–H) qPCR analysis of the indicated genes in NIH3T3 cells expressing YAP5SA and YAP5SA-S94A (E) or NIH3T3 cells treated with 10% FBS for 1 hr after overnight SS (F) or cultured under sparse or confluent conditions in standard culture medium (G) or cultured on soft (1 kPa) to stiff (40 kPa) matrices (H). Kruskal–Wallis test (A), Mann–Whitney U test (B–D), one-way ANOVA (E), and two-tailed Student’s t-test (F–H) were performed to assess statistical significance. These data (A–H) are representative of three independent experiments.
-
Figure 2—figure supplement 2—source data 1
Original data for the statistical analysis in Figure 2—figure supplement 2A–H.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig2-figsupp2-data1-v1.xlsx
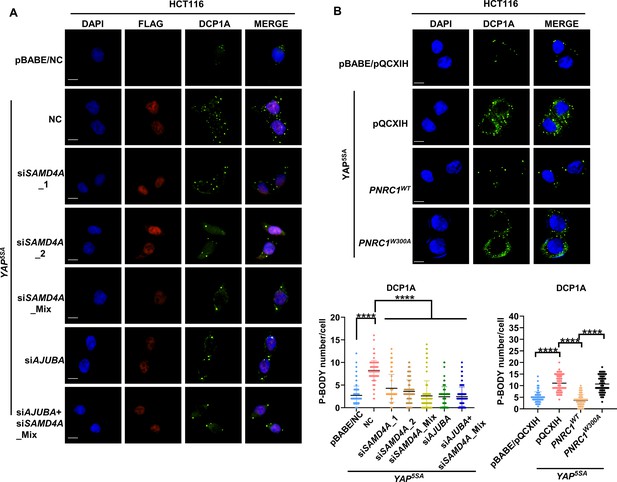
SAMD4A, AJUBA, and PNRC1 mediate the regulatory functions of YAP/TAZ in P-body formation.
(A) Immunofluorescence analysis of DDX6 and DCP1A in HCT116 cells stably expressing YAP5SA and YAP5SA-expressing cells transiently transfected with SMAD4A and AJUBA siRNA. Foci were counted in 100 cells per group. (B) Immunofluorescence analysis of DDX6 and DCP1A in HCT116 cells expressing YAP5SA alone or in combination with PNRC1WT or PNRC1W300A. Kruskal–Wallis test was performed to assess statistical significance. These data (A–B) are representative of three independent experiments.
-
Figure 3—source data 1
Original data for the statistical analysis in Figure 3A and B.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig3-data1-v1.xlsx
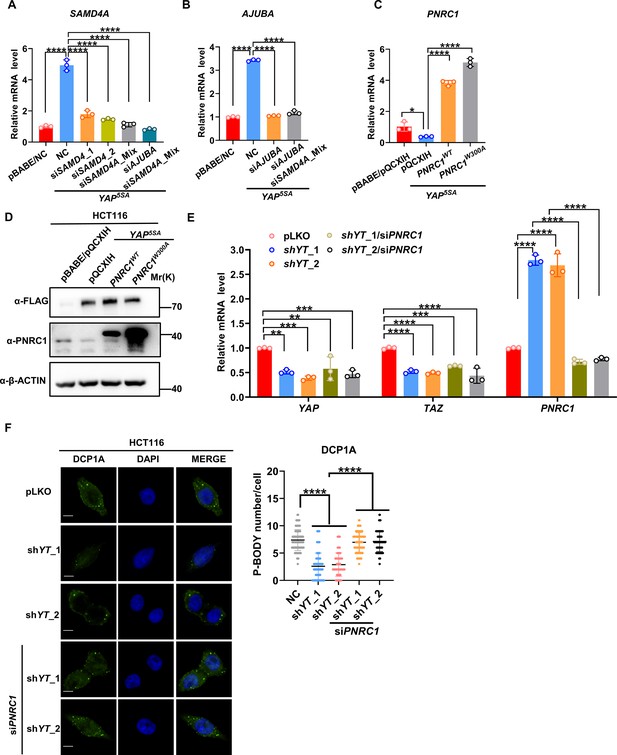
Knockdown of PNRC1 reverses the attenuated P-body formation induced by YAP/TAZ knockdown.
(A, B) qPCR analysis of the mRNA levels of SAMD4A (A) and AJUBA (B) in HCT116 cells stably expressing FLAG-YAP5SA and YAP5SA and transfected with SMAD4A or AJUBA siRNA. n = 3 biologically independent samples per group. (C, D) qPCR (C) and western blot (D) analysis of PNRC1 expression in HCT116 cells stably expressing FLAG-YAP5SA alone or in combination with PNRC1WT or PNRC1W300A. (E) qPCR analysis of the mRNA level of PNRC1/YAP/TAZ in YAP/TAZ knockdown HCT116 cells transfected with PNRC1 siRNA. n = 3 biologically independent samples per group. (F) Immunofluorescence analysis of DCP1A in YAP/TAZ knockdown HCT116 cells transfected with PNRC1 siRNA. Foci were counted in 100 cells per group. One-way ANOVA (A–C, E) and Kruskal–Wallis test (F) were performed to assess statistical significance for qPCR analysis in this figure. The data (F) is representative of two independent experiments.
-
Figure 3—figure supplement 1—source data 1
Original data for the statistical analysis in Figure 3—figure supplement 1A–F.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig3-figsupp1-data1-v1.xlsx
-
Figure 3—figure supplement 1—source data 2
Original file for the western blot analysis in Figure 3—figure supplement 1D.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig3-figsupp1-data2-v1.pdf
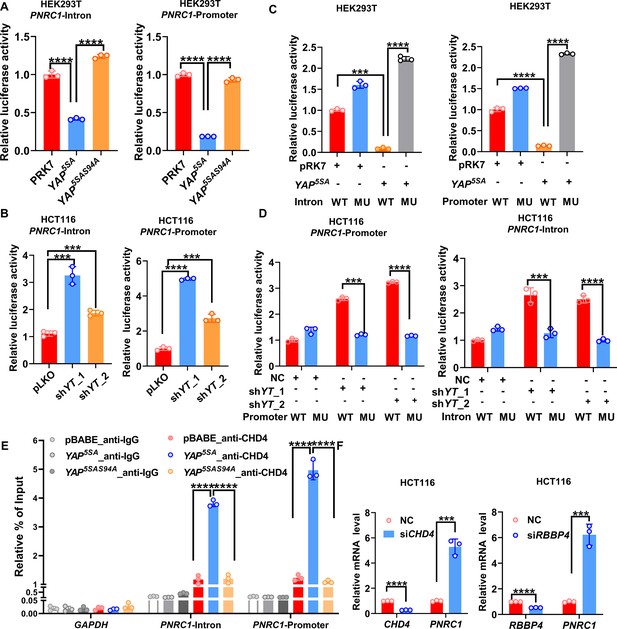
YAP suppresses PNRC1 gene transcription by recruiting the NuRD complex.
(A) Overexpression of YAP5SA but not YAP5SA-S94A decreased the luciferase activity of the PNRC1 promoter and intron reporters. HEK-293T cells were transfected with the indicated FLAG-YAP5SA and YAP5SA-S94A expression plasmids and the PNRC1 promoter or intron luciferase reporter. (B) Knockdown of YAP/TAZ stimulated the luciferase activity of the PNRC1 promoter and intron reporters. The PNRC1 promoter or intron luciferase reporter plasmid and the Renilla luciferase reporter plasmid were co-transfected into HCT116 cells stably expressing pLKO-vec, shYAP/TAZ-1, or shYAP/TAZ-2. (C, D) Luciferase assay of the PNRC1 promoter/intron WT reporters and mutant reporters with TEAD binding motif mutations in HEK-293T cells (C) and HCT116 cells (D). (E) ChIP–qPCR analysis of CHD4 binding to the PNRC1 promoter and intronic regions in control and HCT116 cells stably expressing FLAG-YAP5SA or YAP5SA-S94A. (F) qPCR analysis of PNRC1, CHD4, and RBBP4 in HCT116 cells transfected with the indicated siRNAs. n = 3 biologically independent samples per group. One-way ANOVA (A–E) and two-tailed Student’s t-test (F) were performed to assess statistical significance in this figure. These data (A–F) are representative of two independent experiments.
-
Figure 4—source data 1
Original data for the statistical analysis in Figure 4A–F.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig4-data1-v1.xlsx
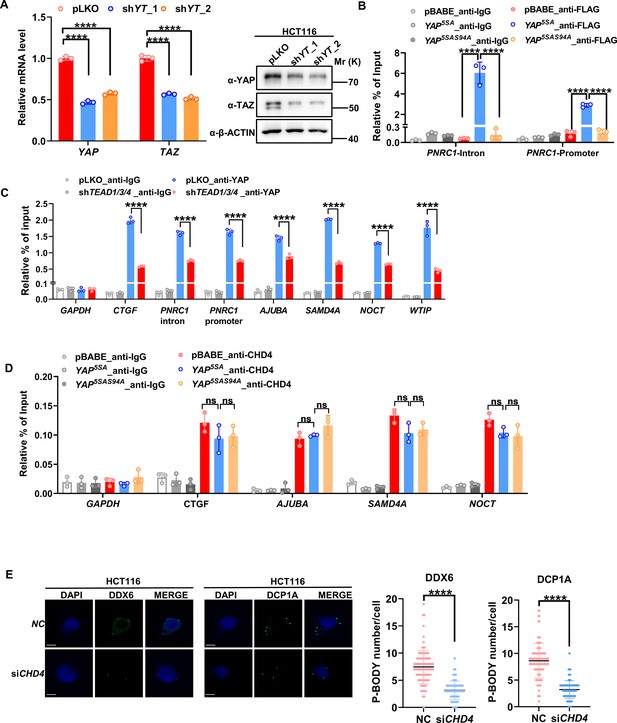
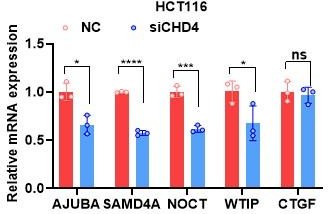
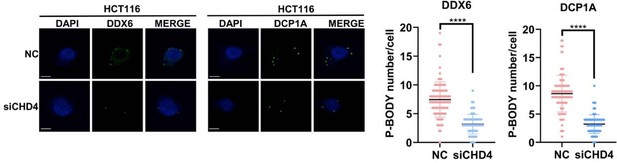
TEADs and CHD4 mediates YAP-dependent inhibition on PNRC1 gene transcription.
(A) qPCR and western blot analysis of the YAP/TAZ knockdown stable HCT116 cells. (B) ChIP–qPCR analysis of FLAG-YAP5SA, YAP5SA-S94A binding to the PNRC1 promoter and intronic regions in control and HCT116 cells stably expressing FLAG-YAP5SA or YAP5SA-S94A. (C) ChIP–qPCR analysis of YAP binding to the genomic locus of the indicated P-body-related genes in control and HCT116 cells with stably knockdown of TEAD1/3/4. (D) ChIP–qPCR analysis of CHD4 binding to the YAP binding sites of indicated genes in control and HCT116 cells stably expressing FLAG-YAP5SA or YAP5SA-S94A. One-way ANOVA was performed to assess statistical significance for qPCR analysis in this figure. These data (B–D) are representative of two independent experiments. (E) Immunofluorescence analysis of DDX6 and DCP1A in HCT116 cells. Cells were transfected with control and CHD4 siRNA for 3 d before Immunofluorescence analysis. Foci were counted in 100 cells per group. Kruskal–Wallis test was performed to assess statistical significance.
-
Figure 4—figure supplement 1—source data 1
Original data for the statistical analysis in Figure 4—figure supplement 1A–E.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig4-figsupp1-data1-v1.xlsx
-
Figure 4—figure supplement 1—source data 2
Original file for the western blot analysis in Figure 4—figure supplement 1A.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig4-figsupp1-data2-v1.pdf
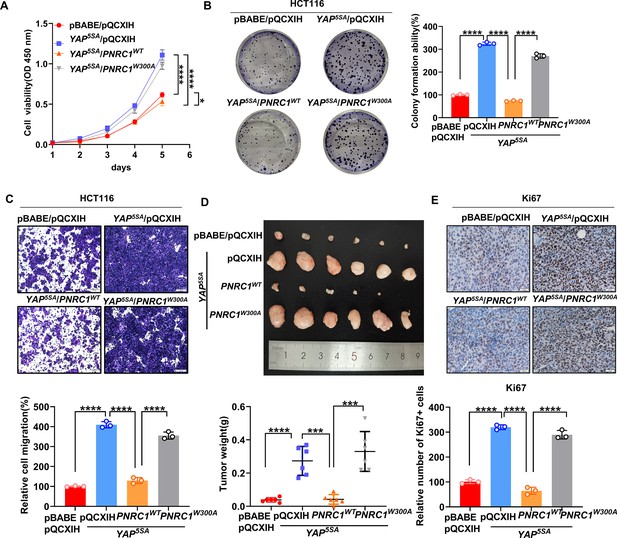
PNRC1 attenuates the oncogenic function of YAP in colorectal cancer (CRC).
(A) CCK8 proliferation assays of HCT116 cells stably expressing YAP5SA alone or in combination with of PNRC1WT or PNRC1W300A. n = 4 biologically independent samples per group. (B, C) Colony formation assay (B) and Transwell assay (C) of HCT116 cells stably expressing YAP5SA alone or in combination with PNRC1WT or PNRC1W300A. n = 3 biologically independent samples per group. (D) Representative images of xenograft tumors formed from HCT116 cells stably expressing YAP5SA alone or in combination with of PNRC1WT or PNRC1W300A (n = 6). (E) Representative images of IHC staining of the proliferation marker Ki67 in xenograft tumors formed from HCT-116 cells stably expressing YAP5SA alone or in combination with PNRC1WT or PNRC1W300A (n = 3). Two-way ANOVA (A) and one-way ANOVA (B–E) were performed to assess statistical significance in this figure. These data (A–C) are representative of two independent experiments.
-
Figure 5—source data 1
Original data for the statistical analysis in Figure 5A–E.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig5-data1-v1.xlsx
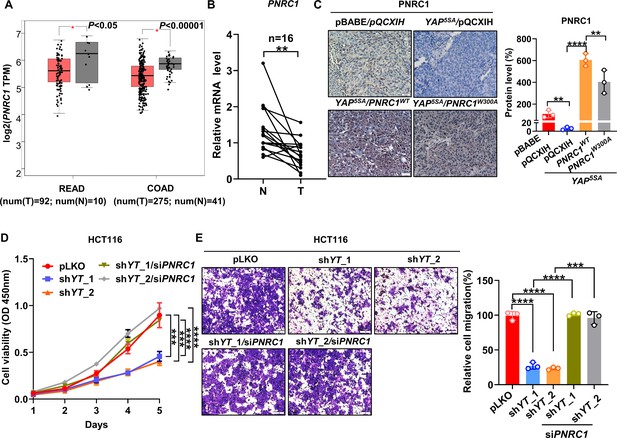
PNRC1 is a tumor suppressor gene in CRC.
(A) Downregulated mRNA expression of PNRC1 in colon cancer (COAD) and rectal cancer (READ) colorectal cancer (CRC) datasets in TCGA. The mRNA levels of PNRC1 were extracted from the GEPIA database. (B) qPCR analysis of the mRNA levels of PNRC1 in 16 paired normal mucosa and colorectal tumor tissues. Paired Student’s t-test was performed to assess statistical significance. (C) Representative images of IHC staining of PNRC1 in xenograft tumors formed from HCT116 cells stably expressing YAP5SA alone or in combination with PNRC1WT or PNRC1W300A (n = 3). (D, E) CCK8 proliferation assay (n = 4) (D) and Transwell migration assay (n = 3) (E) of YAP/TAZ knockdown HCT116 cells transfected with PNRC1 siRNA. One-way ANOVA (A, C, E) and two-way ANOVA (D) were performed to assess statistical significance. These data (D, E) are representative of three independent experiments.
-
Figure 5—figure supplement 1—source data 1
Original data for the statistical analysis in Figure 5—figure supplement 1B–E.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig5-figsupp1-data1-v1.xlsx
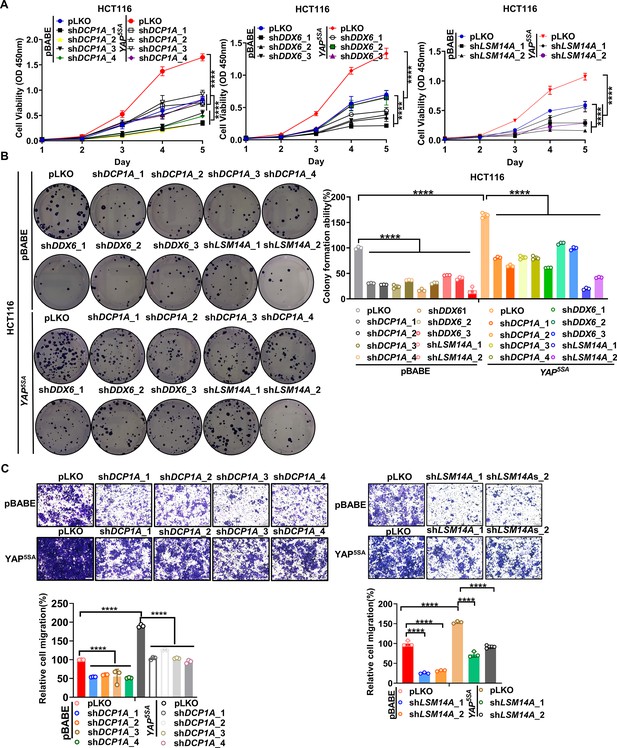
Knockdown of P-body-related core genes suppresses the oncogenic function of YAP in colorectal cancer (CRC).
(A) CCK8 proliferation assays of control HCT116 cells with or without knockdown of DCP1A, LSM14A or DDX6 and HCT116 cells stably expressing YAP5SA with or without knockdown of DCP1A, LSM14A, or DDX6. n = 5 biologically independent samples per group. (B, C) Colony formation assay (B) and Transwell assay (C) of control HCT116 cells with or without knockdown of DCP1A, LSM14A, or DDX6 and HCT116 cells stably expressing YAP5SA with or without knockdown of DCP1A, LSM14A, or DDX6. n = 3 biologically independent samples per group. Two-way ANOVA (A) and one-way ANOVA (B, C) were performed to assess statistical significance in this figure. These data (A–C) are representative of three independent experiments.
-
Figure 6—source data 1
Original data for the statistical analysis in Figure 6A–C.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig6-data1-v1.xlsx
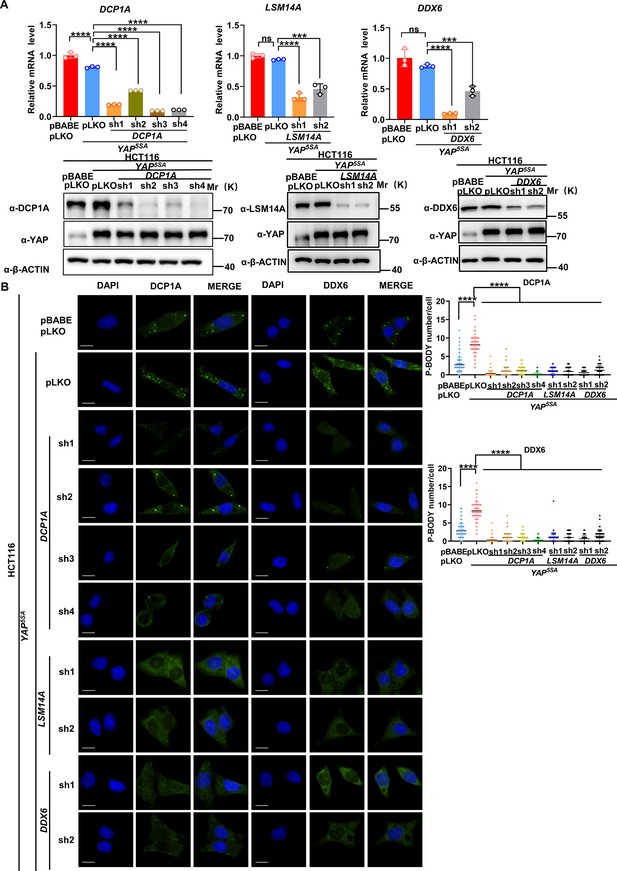
Knockdown efficiency and P-body reduction in DCP1A/LSM14A/DDX6 knockdown HCT116 cells.
(A) qPCR and western blot analysis of the knockdown efficiency of DCP1A, LSM14A, and DDX6 in HCT116 cells stably expressing YAP5SA with or without knockdown of DCP1A, LSM14A, or DDX6. n = 3 biologically independent samples per group. (B) Immunofluorescence analysis of DDX6 and DCP1A in HCT116 cells stably expressing YAP5SA with or without knockdown of DCP1A, LSM14A, or DDX6. Foci were counted in 100 cells per group. One-way ANOVA (A) and Kruskal–Wallis test (B) were performed to assess statistical significance.
-
Figure 6—figure supplement 1—source data 1
Original data for the statistical analysis in Figure 6—figure supplement 1A and B.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig6-figsupp1-data1-v1.xlsx
-
Figure 6—figure supplement 1—source data 2
Original file for the western blot analysis in Figure 6—figure supplement 1A.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig6-figsupp1-data2-v1.pdf
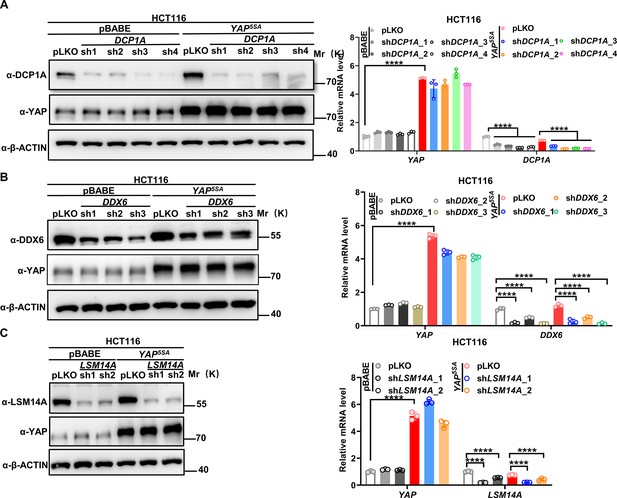
Confirmation of DCP1A/ LSM14A/DDX6 knockdown in YAP5SA-expressing HCT116 cells.
(A–C) qPCR and western blot analysis of the knockdown efficiency of DCP1A (A), DDX6 (B), and LSM14A (C) in control HCT116 cells with or without knockdown of DCP1A, LSM14A, or DDX6 and stable YAP5SA overexpression HCT116 cells with or without knockdown of DCP1A, LSM14A, or DDX6. n = 3 biologically independent samples per group. One-way ANOVA was performed to assess statistical significance.
-
Figure 6—figure supplement 2—source data 1
Original data for the statistical analysis in Figure 6—figure supplement 2A–C.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig6-figsupp2-data1-v1.xlsx
-
Figure 6—figure supplement 2—source data 2
Original file for the western blot analysis in Figure 6—figure supplement 2A–C.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig6-figsupp2-data2-v1.pdf
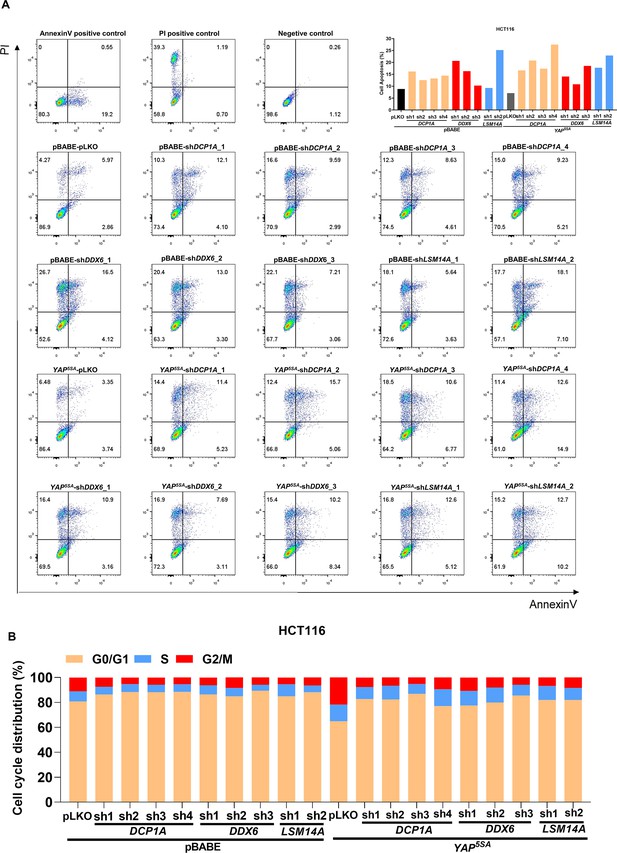
Knockdown of DCP1A, LSM14A and DDX6 downregulates cell mitosis and increases cell apoptosis in YAP5SA-expressing HCT116 cells.
(A, B) Cell apoptosis (A) and cell cycle (B) analysis of the control HCT116 cells with or without knockdown of DCP1A, LSM14A, or DDX6 and stable YAP5SA overexpression HCT116 cells with or without knockdown of DCP1A, LSM14A, or DDX6.
-
Figure 6—figure supplement 3—source data 1
Original data for the bar plot in Figure 6—figure supplement 3A and B.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig6-figsupp3-data1-v1.xlsx
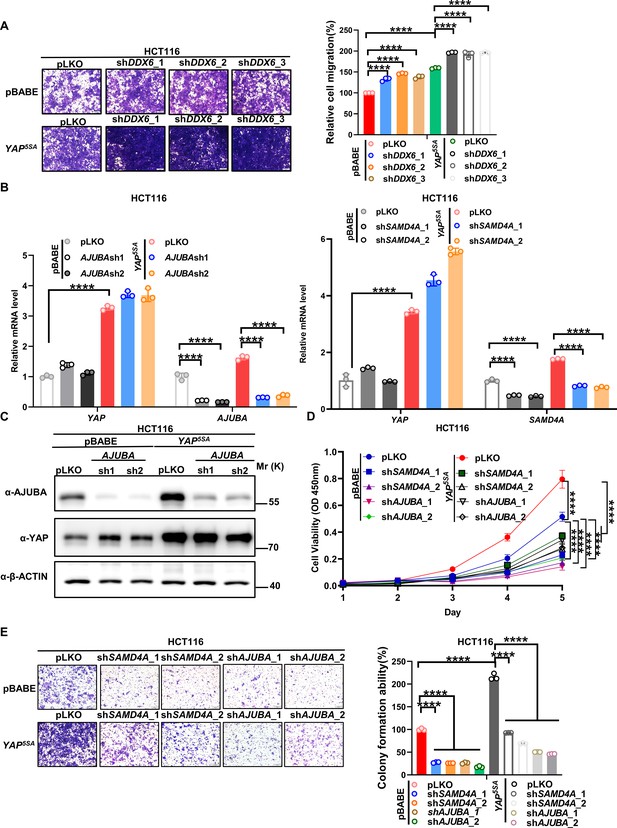
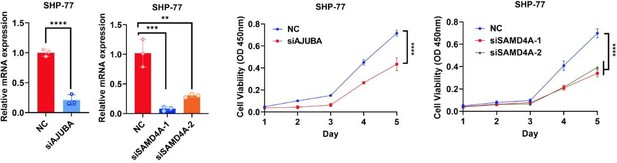
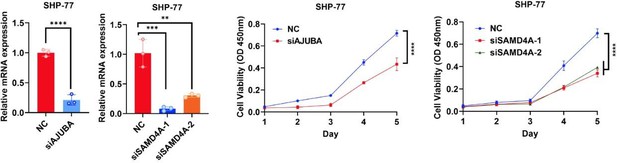
Knockdown of AJUBA or SAMD4A attenuates the oncogenic function of YAP in HCT116 cells.
(A) Transwell assay of control HCT116 cells with or without knockdown of DDX6 and HCT116 cells stably expressing YAP5SA with or without knockdown of DDX6. n = 3 biologically independent samples per group. (B, C) qPCR (B) and western blot (C) analysis of the knockdown efficiency of AJUBA and SAMD4A in control HCT116 cells with or without knockdown of AJUBA or SAMD4A and stable YAP5SA overexpression HCT116 cells with or without knockdown of AJUBA or SAMD4A. n = 3 biologically independent samples per group. (D, E) CCK8 proliferation (n = 5) (D) and Transwell (E) assays of control HCT116 cells with or without knockdown of AJUBA or SAMD4A and stable YAP5SA overexpression HCT116 cells with or without knockdown of AJUBA or SAMD4A. One-way ANOVA (A, B, E) and two-way ANOVA (D) were performed to assess statistical significance in this figure. These data (A, D, E) are representative of three independent experiments.
-
Figure 6—figure supplement 4—source data 1
Original data for the statistical analysis in Figure 6—figure supplement 4A–D.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig6-figsupp4-data1-v1.xlsx
-
Figure 6—figure supplement 4—source data 2
Original file for the western blot analysis in Figure 6—figure supplement 4C.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig6-figsupp4-data2-v1.pdf
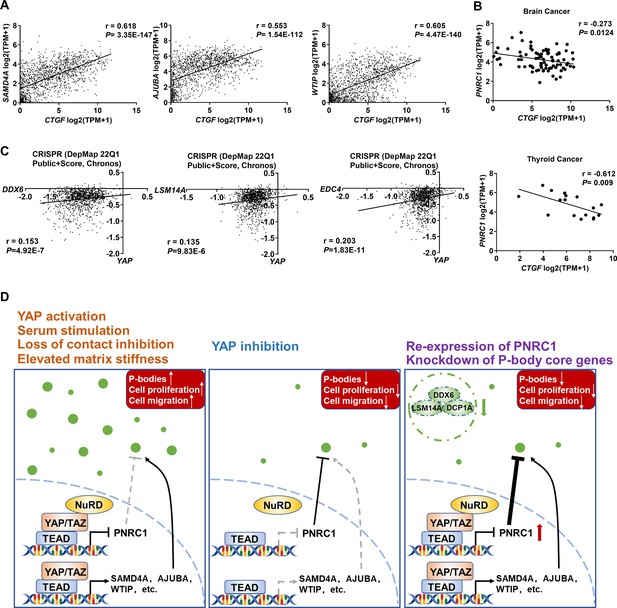
DepMap analysis reveals the co-dependencies of YAP/TEAD and P-body core genes in pancancer CRISPR screens.
(A) Positive correlations between the mRNA levels of CTGF and SAMD4A/AJUBA/WTIP in 1393 cancer cell lines. (B) Negative correlation between the mRNA levels of CTGF and PNRC1 in brain cancer cell lines (n = 83) and thyroid cancer cell lines (n = 17). (C) Positive correlations between the dependency scores of YAP and DDX6/LSM14A/EDC4 in 1070 cancer cell lines. The Chronos dependency scores were extracted from the DepMap database. The negative Chronos score indicates decreased cell proliferation upon gene knockout. Pearson correlation analysis was used to assess statistical significance. (D) In response to serum stimulation or under loss of contact inhibition or reduced ECM stiffness, activation of YAP enhances the P-body formation to promote colorectal cancer (CRC) cell proliferation and migration. Disruption of P-bodies by overexpression of the tumor suppressor gene PNRC1 or knockdown of P-body core genes could attenuate the cell proliferation and migration induced by activation of YAP in CRC cells.
-
Figure 7—source data 1
Original data for the statistical analysis in Figure 7A–C.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig7-data1-v1.xlsx
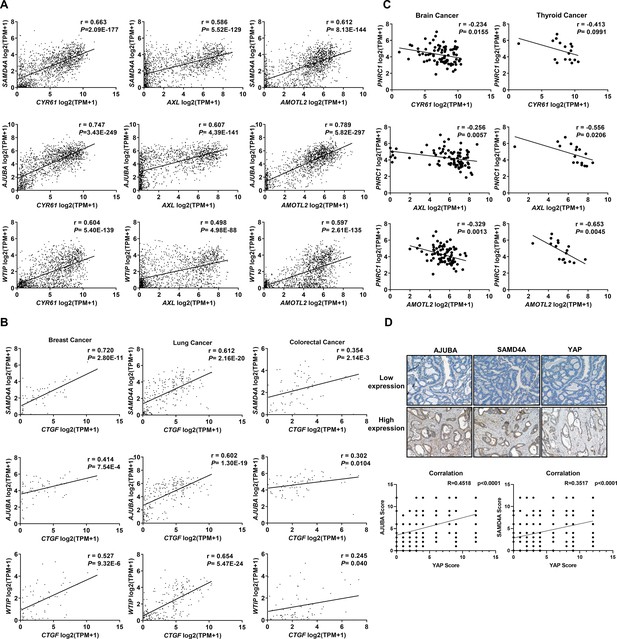
Correlation analysis of the expression of YAP canonical target genes and YAP-related P-body genes.
(A) Positive correlations between the mRNA levels of YAP target genes (CYR61, AXL, and AMOTL2) and SAMD4A/AJUBA/WTIP in 1393 cancer cell lines. (B) Positive correlations between the mRNA levels of YAP target genes (CTGF) and SAMD4A/AJUBA/WTIP in brain cancer (n = 83), lung cancer (n = 207), and colorectal cancer (n = 71) cell lines. (C) Negative correlations between the mRNA levels of YAP target genes (CYR61, AXL, and AMOTL2) and PNRC1 in brain cancer cell lines (n = 83) and thyroid cancer cell lines (n = 17). (D) IHC analysis of AJUBA, SAMD4A, and YAP protein expression in colorectal cancer tissues (n = 294). Pearson (A–C) and Spearman (D) correlation analysis was used to assess statistical significance.
-
Figure 7—figure supplement 1—source data 1
Original data for the statistical analysis in Figure 7—figure supplement 1A–D.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig7-figsupp1-data1-v1.xlsx
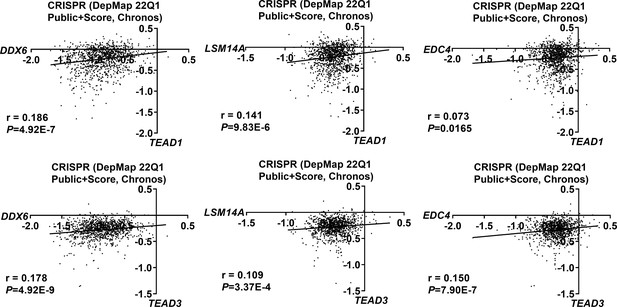
The TEAD1/3 dependency scores are positively correlated with the EDC4/DDX6/LSM14A scores.
Positive correlations between the dependency scores of TEAD1/3 and DDX6/LSM14A/EDC4 in 1070 cancer cell lines. The Chronos dependency scores were extracted from the DepMap database. The negative Chronos score indicates decreased cell proliferation upon gene knockout. Pearson correlation analysis was used to assess statistical significance.
-
Figure 7—figure supplement 2—source data 1
Original data for the statistical analysis in Figure 7—figure supplement 2A.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig7-figsupp2-data1-v1.xlsx
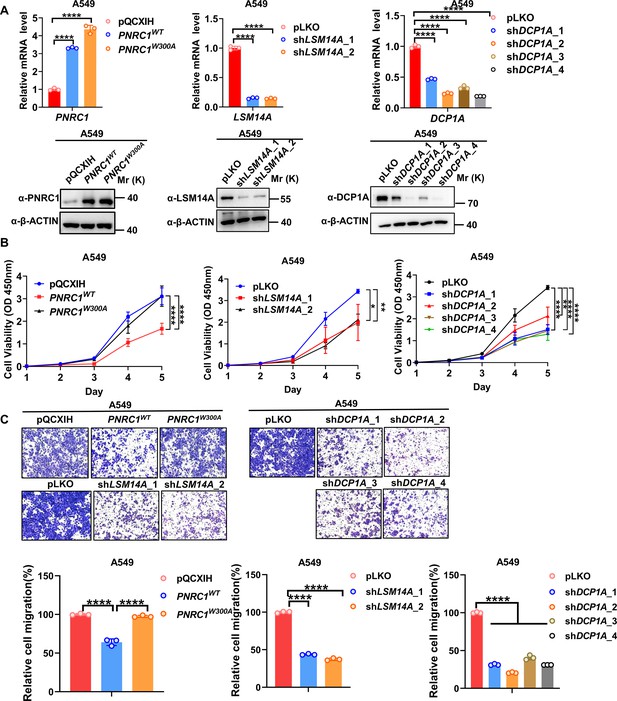
Knockdown of DCP1A/LSM14A and overexpression of PNRC1 suppress both cell proliferation and cell migration in A549 lung cancer cells.
(A) qPCR and western blot analysis of the knockdown efficiency of DCP1A / LSM14A or PNRC1 overexpression in A549 cells stably expressing PNRC1WT/PNRC1W300A or with knockdown of DCP1A and LSM14A. (B, C) CCK8 proliferation (n = 5 for PNRC1 OE, n = 3 for knockdown of DCP1A and LSM14A) (B) and Transwell (C) assays of A549 cells stably expressing PNRC1WT/PNRC1W300A or with knockdown of DCP1A and LSM14A. Two-way ANOVA (B) and one-way ANOVA (C) were performed to assess statistical significance in this figure. These data (B, C) are representative of two independent experiment.
-
Figure 7—figure supplement 3—source data 1
Original data for the statistical analysis in Figure 7—figure supplement 3A–C.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig7-figsupp3-data1-v1.xlsx
-
Figure 7—figure supplement 3—source data 2
Original file for the western blot analysis in Figure 7—figure supplement 3A.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig7-figsupp3-data2-v1.pdf
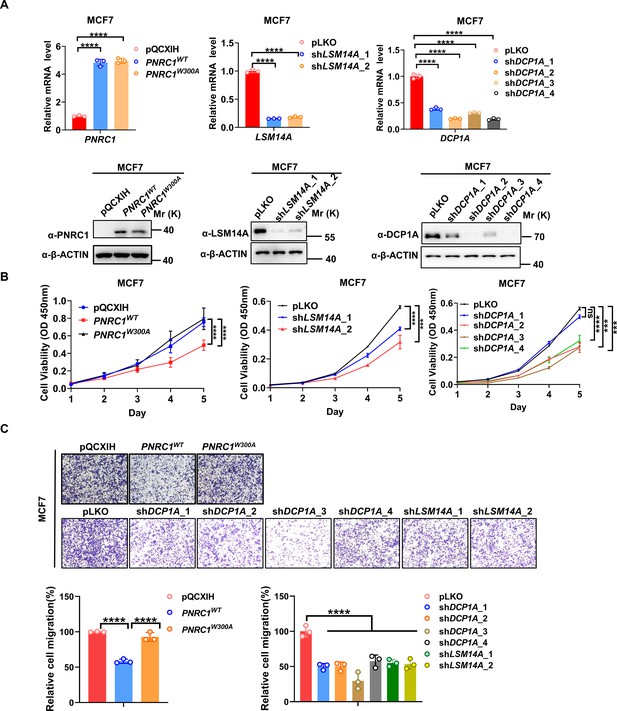
Knockdown of DCP1A/LSM14A and overexpression of PNRC1 suppress both cell proliferation and cell migration in MCF7 breast cancer cells.
(A) qPCR and western blot analysis of the knockdown efficiency of DCP1A / LSM14A or PNRC1 overexpression in MCF7 cells stably expressing PNRC1WT/PNRC1W300A or with knockdown of DCP1A and LSM14A. (B, C) CCK8 proliferation (n = 6 for PNRC1 OE, n = 3 for knockdown of DCP1A and LSM14A) (B) and transwell (C) assays of MCF7 cells stably expressing PNRC1WT/PNRC1W300A or with knockdown of DCP1A and LSM14A. Two-way ANOVA (B) and one-way ANOVA (C) were performed to assess statistical significance in this figure. These data (B, C) are representative of 2 independent experiments.
-
Figure 7—figure supplement 4—source data 1
Original data for the statistical analysis in Figure 7—figure supplement 4A–C.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig7-figsupp4-data1-v1.xlsx
-
Figure 7—figure supplement 4—source data 2
Original file for the western blot analysis in Figure 7—figure supplement 4A.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig7-figsupp4-data2-v1.pdf
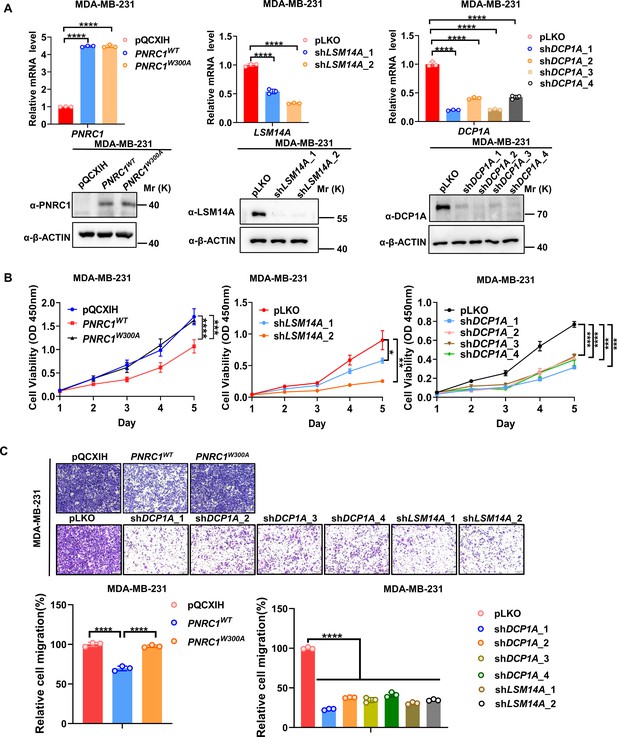
Knockdown of DCP1A/LSM14A and overexpression of PNRC1 suppress both cell proliferation and cell migration in MDA-MB-231 breast cancer cells.
(A) qPCR and western blot analysis of the knockdown efficiency of DCP1A / LSM14A or PNRC1 overexpression in MDA-MB-231 cells stably expressing PNRC1WT/PNRC1W300A or with knockdown of DCP1A and LSM14A. (B, C) CCK8 proliferation (n = 6 for PNRC1 OE, n = 3 for knockdown of DCP1A and LSM14A) (B) and transwell (C) assays of MDA-MB-231 cells stably expressing PNRC1WT/PNRC1W300A or with knockdown of DCP1A and LSM14A. Two-way ANOVA (B) and one-way ANOVA (C) were performed to assess statistical significance in this figure. These data (B, C) are representative of 2 independent experiments.
-
Figure 7—figure supplement 5—source data 1
Original data for the statistical analysis in Figure 7—figure supplement 5A–C.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig7-figsupp5-data1-v1.xlsx
-
Figure 7—figure supplement 5—source data 2
Original file for the western blot analysis in Figure 7—figure supplement 5A.
- https://cdn.elifesciences.org/articles/88573/elife-88573-fig7-figsupp5-data2-v1.pdf
Additional files
-
Supplementary file 1
The expression of P-body genes in YAP/TAZ knockdown HCT116 cells.
The differentially expressed genes in YAP/TAZ knockdown HCT116 cells were analyzed by RNA-seq (GSE176475). The expression levels of genes that were annotated as related to P-bodies are included in this supplementary file.
- https://cdn.elifesciences.org/articles/88573/elife-88573-supp1-v1.xlsx
-
Supplementary file 2
Top 100 correlation for EDC4 in CRISPR (DepMap 22Q1 Public Score, Chronos).
The gene dependency scores of the top 100 genes that are correlated with EDC4 were downloaded from the Cancer Dependency Map (DepMap) database.
- https://cdn.elifesciences.org/articles/88573/elife-88573-supp2-v1.xlsx
-
Supplementary file 3
siRNA, shRNA, and primers used in this study.
The sequences of the siRNA, shRNA, and primers used in this study are included in this supplementary file.
- https://cdn.elifesciences.org/articles/88573/elife-88573-supp3-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/88573/elife-88573-mdarchecklist1-v1.docx