What AlphaFold tells us about cohesin’s retention on and release from chromosomes
Figures
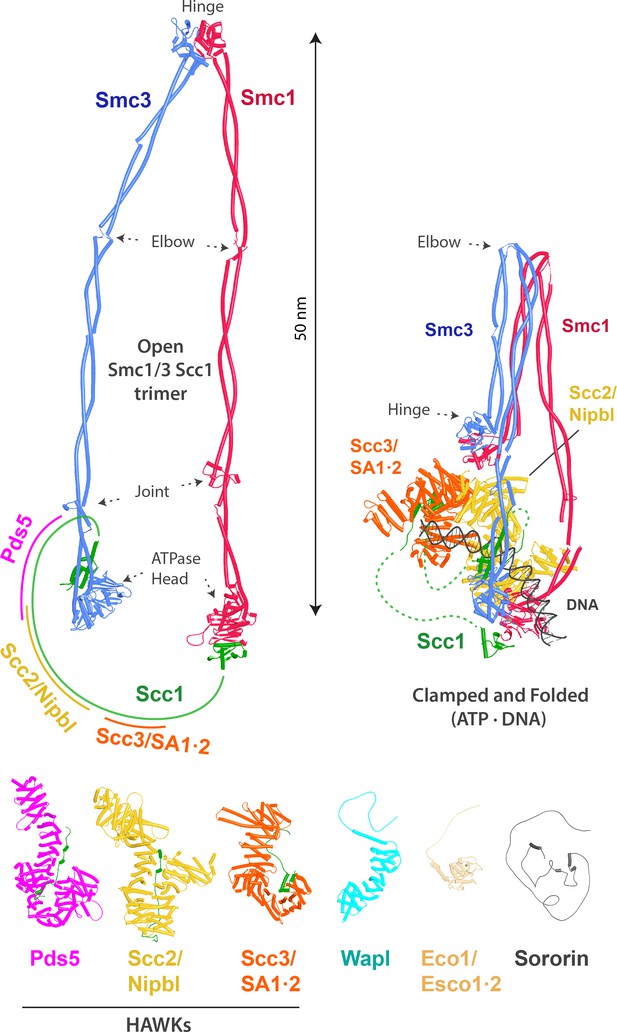
Overview of cohesin, its subunits, and regulatory proteins.
Top left: cohesin’s trimeric Smc1/Smc3/Scc1 ring. The approximate HAWK binding sites on the kleisin Scc1 are indicated. Top right: cohesin in the DNA-clamped and folded state (hybrid model combining PDBs 6WG3, 6ZZ6, and 7OGT). Bottom: cohesin’s HAWKs and regulatory proteins (AlphaFold 2 [AF] predictions).
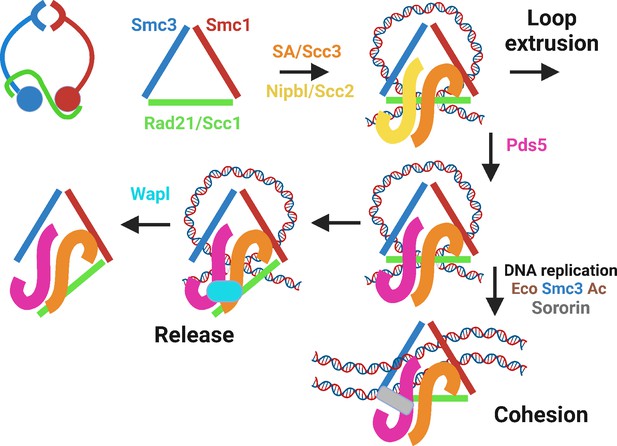
Cohesin’s trimeric ring (top left), represented throughout the rest of the figure as a triangle, associates with DNA loops when occupied by SA/Scc3 and Nipbl/Scc2.
Nipbl/Scc2 stimulates ATP hydrolysis, leading to DNA translocation and loop extrusion. This is halted by Nipbl’s replacement by Pds5, creating a complex that can bind Wapl and detach from chromatin, in a process that depends on a pair of lysine residues (yeast: K112 and K113; humans: K105 and K106) on Smc3’s ATPase head, whose acetylation by Eco1 (yeast) or Esco1/2 in vertebrates blocks release. Acetylated complexes containing Pds5 either form stable loops between CTCF sites or, during DNA replication, generate cohesion between sister DNAs, whose maintenance (in animal and plant cells) requires Sororin (as well as continued acetylation) to prevent Wapl-mediated release.
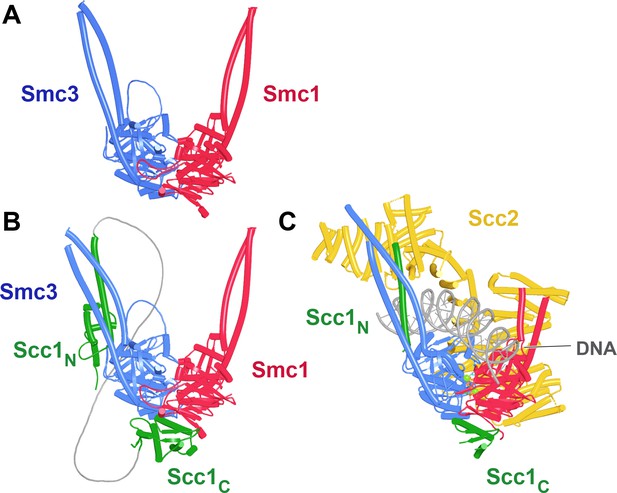
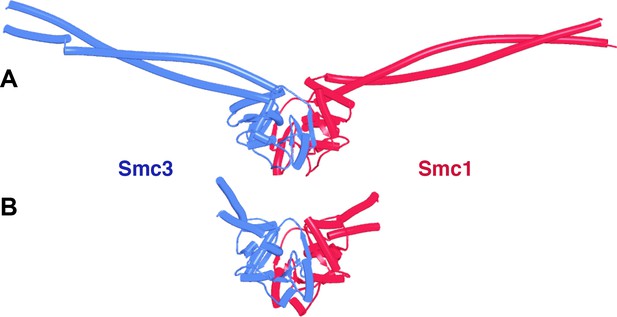
Enagagement of Smc1 and Smc3 heads in the absence and presence of Scc1 and Scc2.
(A) AlphaFold 2 multimer (AF) prediction of yeast Smc1 and Smc3 ATPase heads with associated coiled coils up to, but not including their joints. The actual predictions and sequences used are available in folder f1 at https://doi.org/10.6084%20m9.figshare.22567318.v2. (B) As (A), but including Scc1’s N- and CTD connected by a GS linker (f2). (C) Cryo-EM structure of DNA clamped on top of engaged Smc1 and Smc3 heads by Scc2 (PDB 6ZZ6).
Folding of Smc1 and Smc3 coiled coils is accompanied by their juxtaposition.
(A) AlphaFold 2 (AF) prediction for yeast Smc1 and Smc3 lacking their ATPase domains (f3). (B) As (A), but human Smc1 and Smc3 (f4). (C) Cryo-EM structure of yeast Smc1 and Smc3 hinge and associated coiled coils, PDB 7OGT.
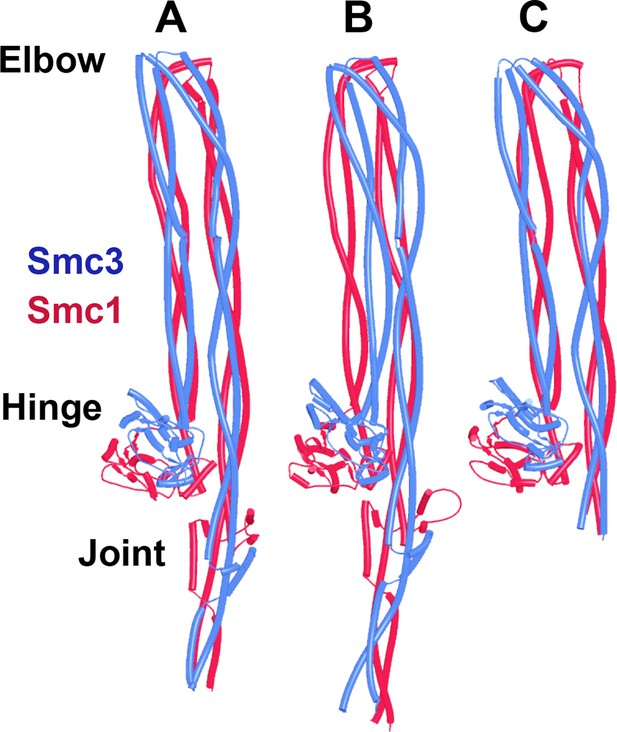
Juxtaposition of Smc1/3 coiled coils requires segments below the elbow.
(A) AlphaFold 2 (AF) prediction for yeast Smc1 and Smc3 hinge domains with short associated coiled coils (f5). (B) Crystal structure of T. maritima hinge associated with short coiled coils, PDB 1GXL.
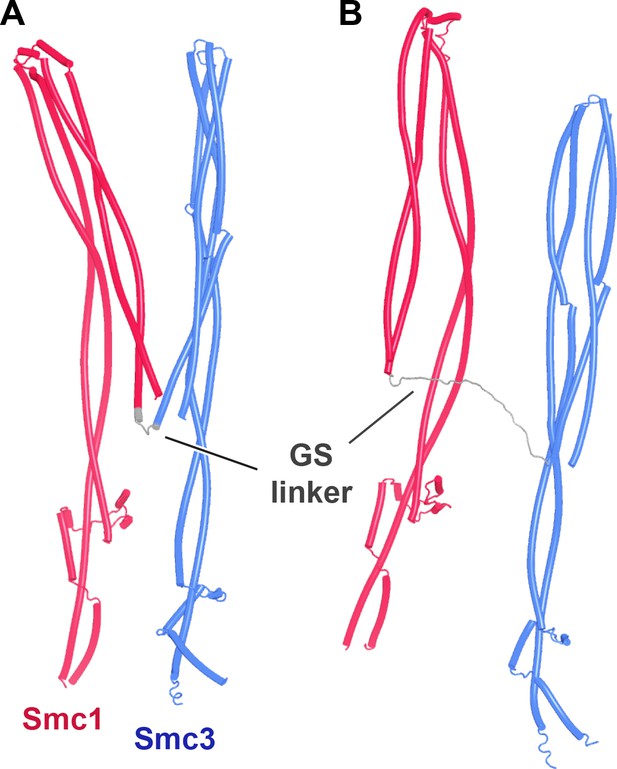
AlphaFold 2 (AF) predictions for yeast Smc1 and Smc3 coiled coils including their elbow and joints, but joined by GSx4 (A, f6) and GSx15 (B, f7) linkers instead of their hinges.
AlphaFold 2 (AF) predictions of Smc1 and Smc3 joints with short associated coiled coils.
(A) (f8) and (B) (f9), yeast.
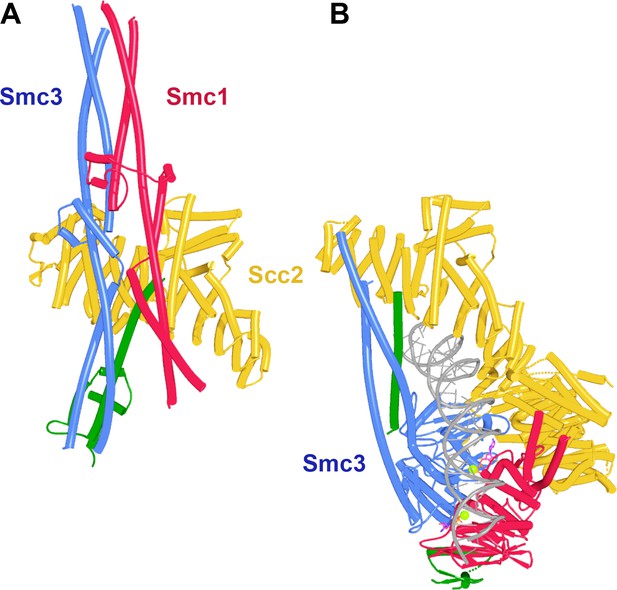
Scc2 binding does not per se disrupt junction of Smc1 and Smc3 joints.
(A) AlphaFold 2 (AF) prediction for yeast Smc1 and Smc3 joints with associated coiled coils together with Scc1 and Scc2’s NTDs (f12). Chains in the PAEs from f12 are Scc2 (A), Smc3 (B), Smc3 (C), Scc1 (D), Smc1 (E), and Smc1 (F). (B) Cryo-EM structure of DNA clamped by Scc2 on top of engaged Smc1 and Smc3 ATPase, PDB 6ZZ6.
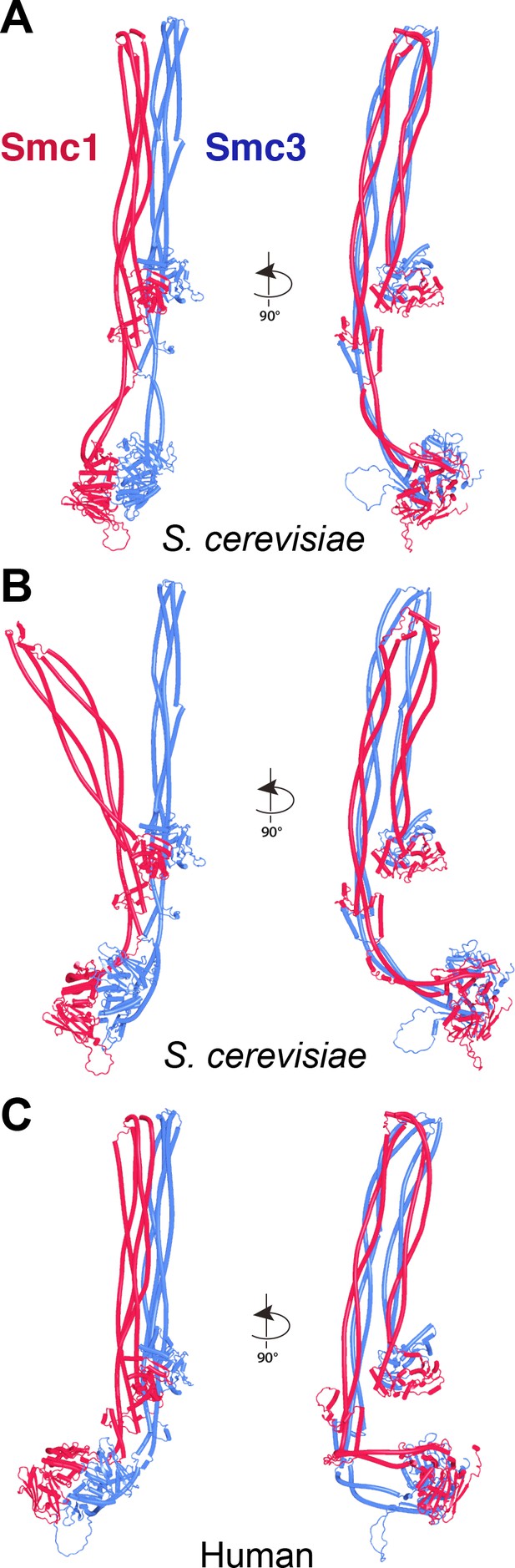
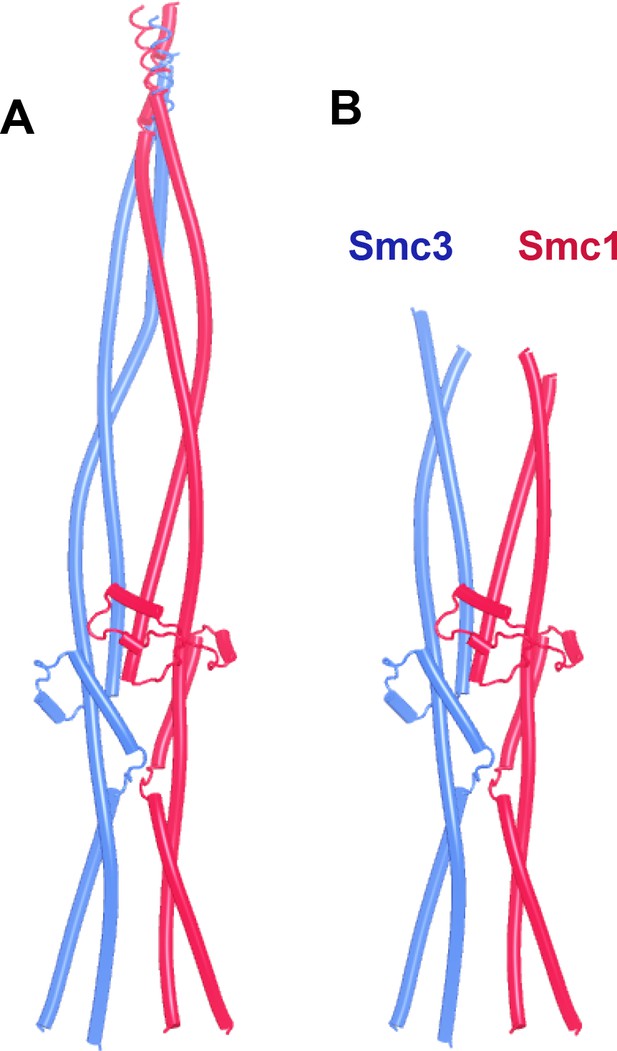
AlphaFold 2 (AF) predictions for full-length yeast (A, B, f13) and human (C, f14) Smc1/Smc3 heterodimers.
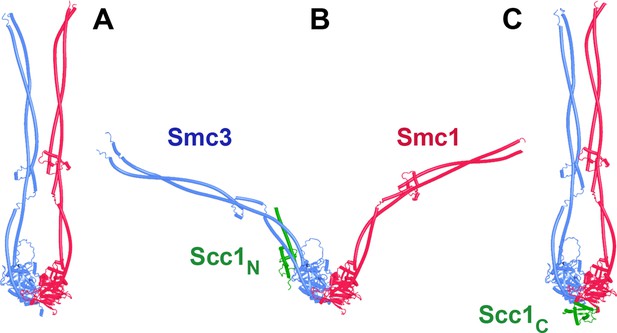
AlphaFold 2 (AF) predictions of yeast Smc1 and Smc3 ATPases with coiled coils extending to the elbow but not beyond, without Scc1 (A, f15), with Scc1’s NTD (B, f16), and with Scc1’s CTD (C, f17).
Chains in the PAEs from f16 are Scc1 (A), Smc1 (B), Smc1 (C), Smc3 (D), and Smc3 (E).
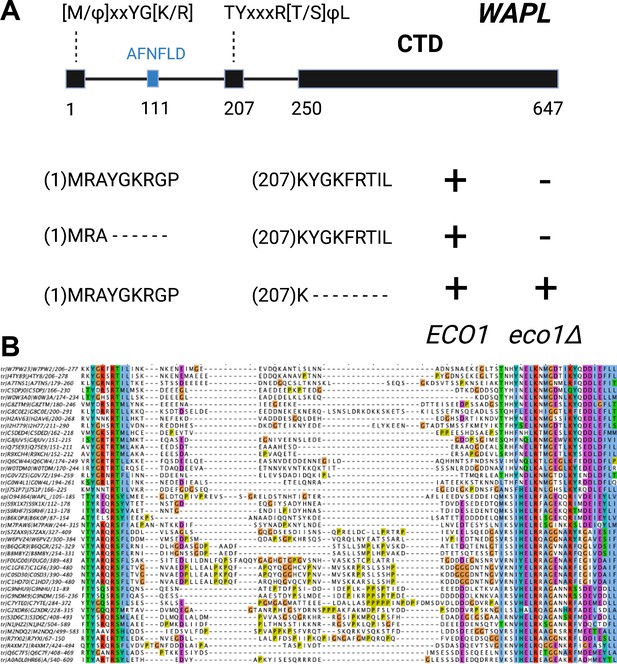
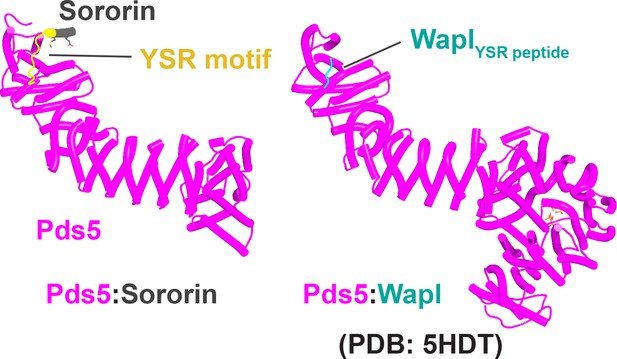
The role and conservation of a short motif within the N-terminal region of fungal Wapl proteins.
(A) Deletion of Wapl’s N-terminal [M/Φ]xxYG[K/R] motif from S. cerevisiae (waplΔ4–9) does not suppress the lethality of eco1 mutations while deletion of its more C-terminal TYxxxR[T/S]ΦL motif (waplΔ208–215) does so. Both deletions were initially created within a version of the WAPL gene tagged with EGFP (KN18714) and their ability to suppress the lethality of eco1Δ determined by crossing to a waplΔ eco1Δ strain (strain KN16432) followed by tetrad analysis. (B) A multiple amino acid sequence alignment of fungal Wapl sequences, showing the TYxxxR[T/S]ΦL motif on the left and the first highly conserved alpha helix within Wapl’s CTD on the right. Yeast Wapl is the top sequence (W7PW2).
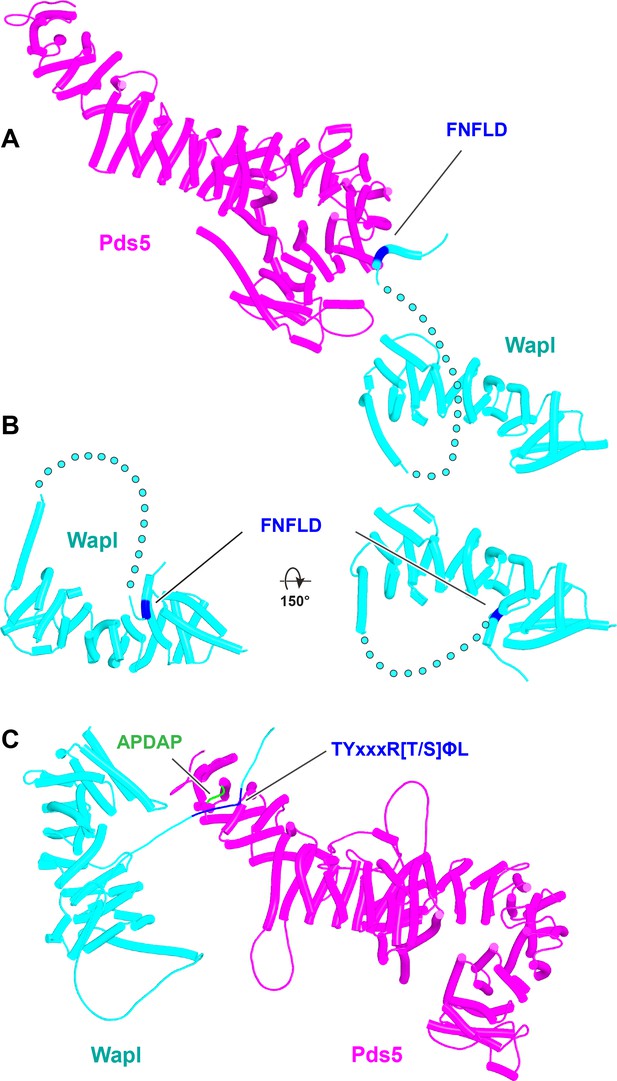
Interactions between motifs within Wapl's N-terminal sequences and Pds5.
(A) AlphaFold 2 (AF) prediction for yeast Wapl’s association with Pds5 (f19A). (B) AF prediction of yeast Wapl alone (AF-Q99359-F1, AlphaFold Protein Structure Database). (C) AF predicts interaction between Wapl’s TYxxxR[T/S]ΦL motif and Pds5’s APDAP loop in Neurospora crassa (f19B).
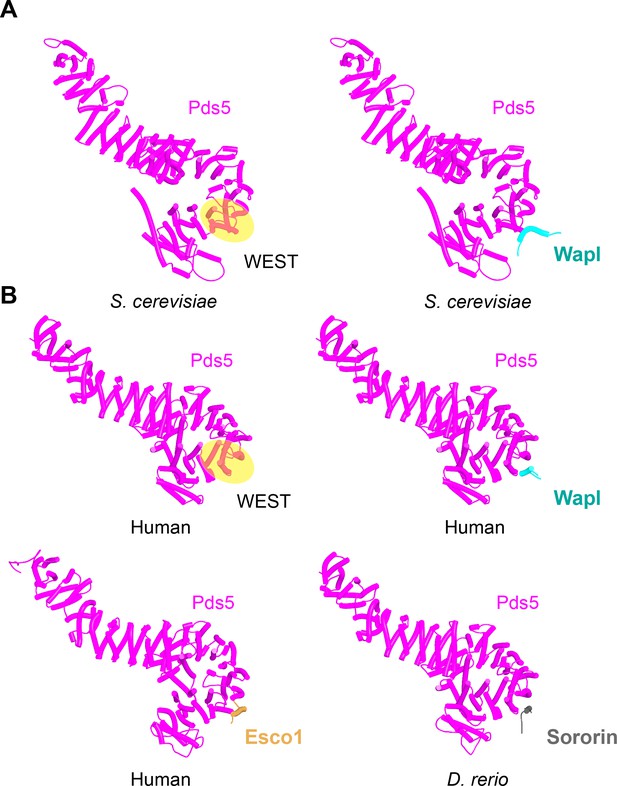
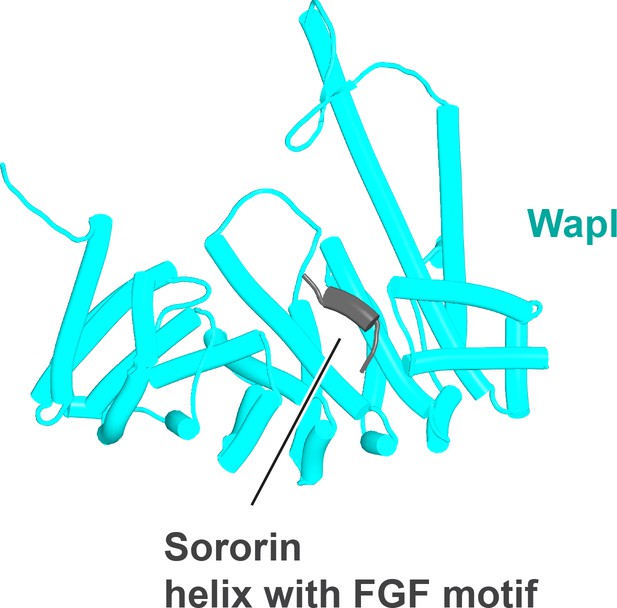
Interaction between a trough in Pds5’s CTD called ‘WEST’ and an FNFLD motif within the N-terminal part of yeast Wapl (A, left and right, f19), FGF motifs within human Wapl (B, upper left and right, f20) and D. rerio Sororin (B, lower right, f21), and a highly conserved, partly helical, ILxLCEEIAGEIESD motif within human Esco1 (B, lower left, f22).
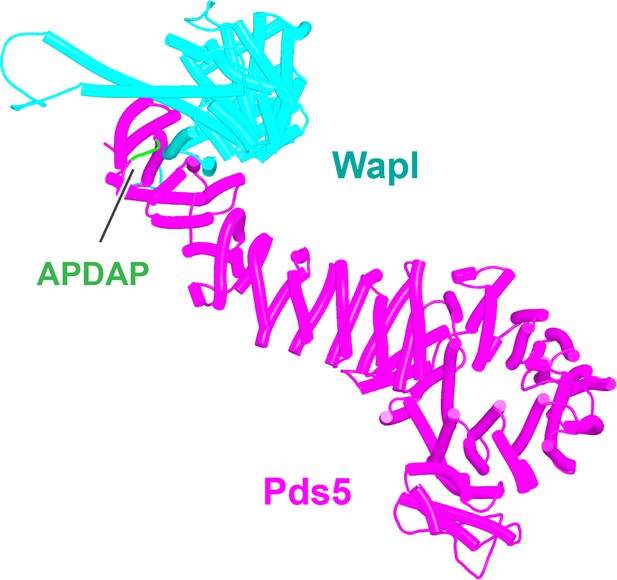
AlphaFold 2 (AF) prediction for the interaction between Pds5A and the N-terminal half of Wapl’s highly conserved CTD (f23).
The same interaction was observed with both Pds5A and B.
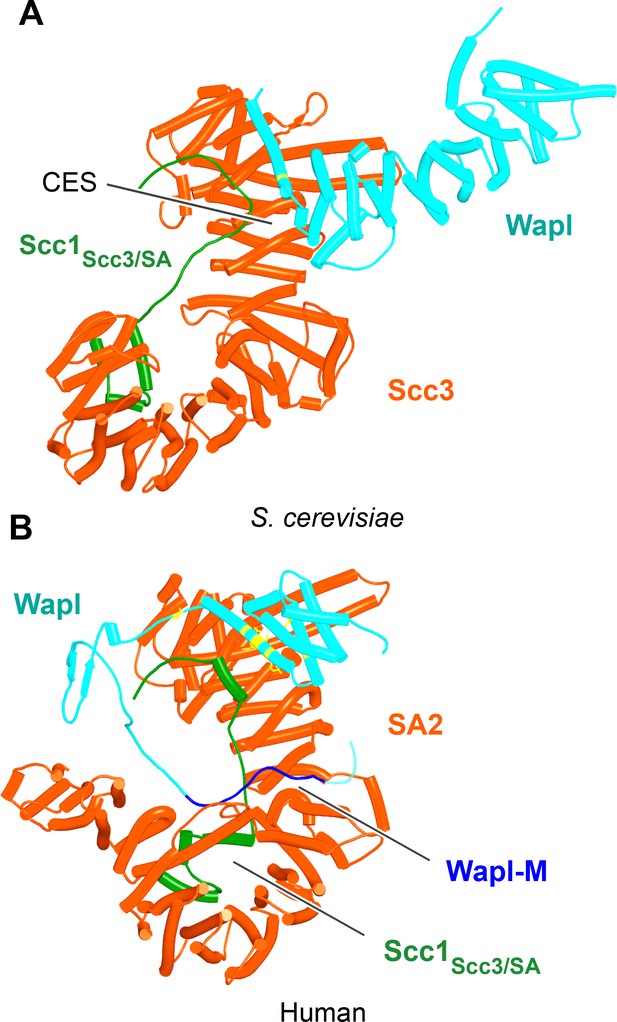
Interaction of Wapl's CTD with Scc3 and SA proteins.
(A) AlphaFold 2 (AF) prediction for the interaction between Scc3/Scc1 and the CTD of yeast Wapl (f24). Chains in the PAEs from f24 are Scc3 (A), Scc1 (B), and Wapl (C). (B) AF prediction for the interaction between SA2/Scc1 and a fragment of human Wapl containing its N-terminal sequences as well as the N-terminal half of its helical CTD (f25). Chains in the PAEs from f25B are Scc1 (A), SA2 (B), and Wapl (C). Mutations that abolish releasing activity (RA) are marked in yellow: E653, D656, D657, L676, and K684.
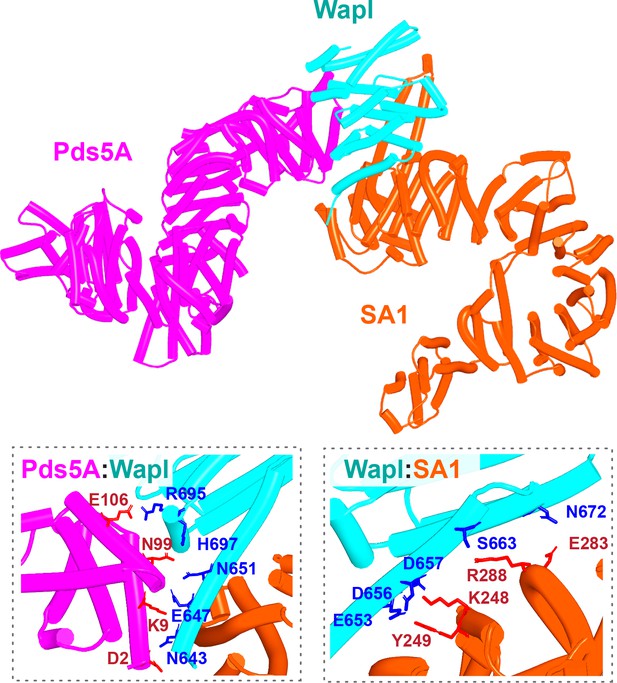
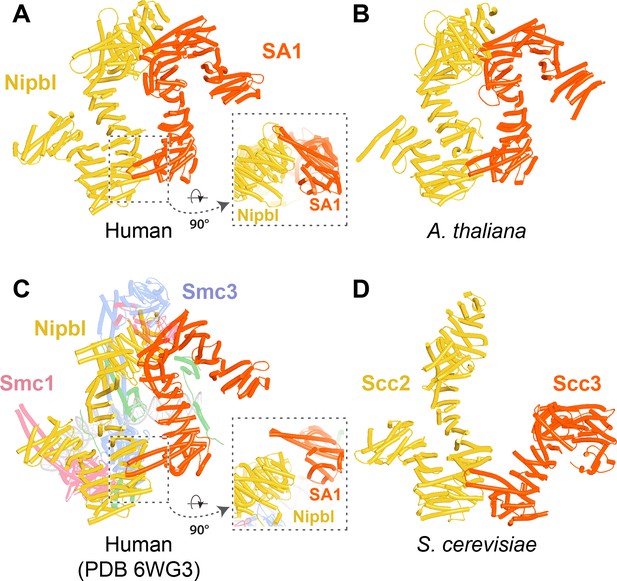
AlphaFold 2 (AF) prediction of a complex between human Pds5A, SA1, and a fragment of Wapl’s CTD (f27).
Chains in the PAEs from f27 are Wapl (A), SA1 (B), and Pds5A (C). Pds5 and SA1 were not predicted to interact in the absence of Wapl (f26).
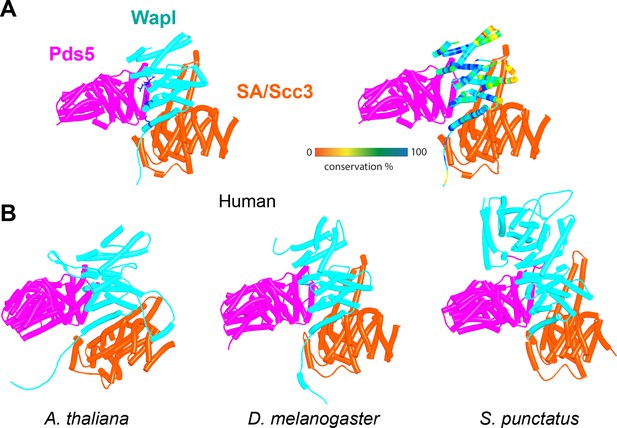
AlphaFold 2 (AF) predictions for complexes between N-terminal fragments of Pds5 and SA/Scc3 and the N-terminal half of Wapl’s CTD for humans (A, f28), A. thaliana (B, f29), D. melanogaster (f30), and S. punctatus (f31).
Also shown is Wapl’s conservation mapped on the human complex (A, right). Chains in the PAEs from f28 are Pds5 (A), Wapl (B), and SA (C). Chains in the PAEs from f29 are Scc3 (A), Pds5 (B), and Wapl (C). Chains in the PAEs from f31 are Pds5 (A), Scc3 (B), and Wapl (C).
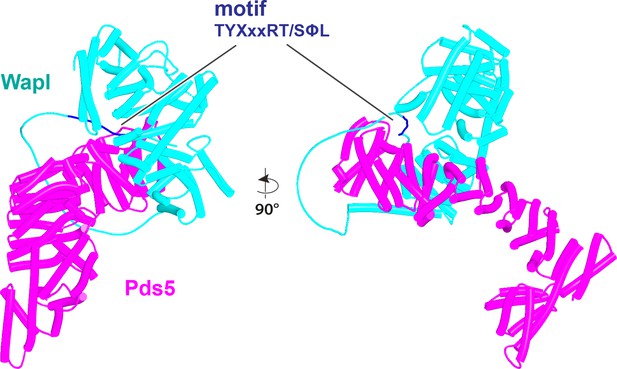
AlphaFold 2 (AF) prediction for the interaction between the N-terminal half of S. punctatus Pds5 and a fragment of Wapl containing its TYxxxRT/SΦL motif connected to its CTD (f19E).
Both the motif and the CTD bind to Pds5. Chains in the PAEs from f19E are Wapl (A) and Pds5 (B).
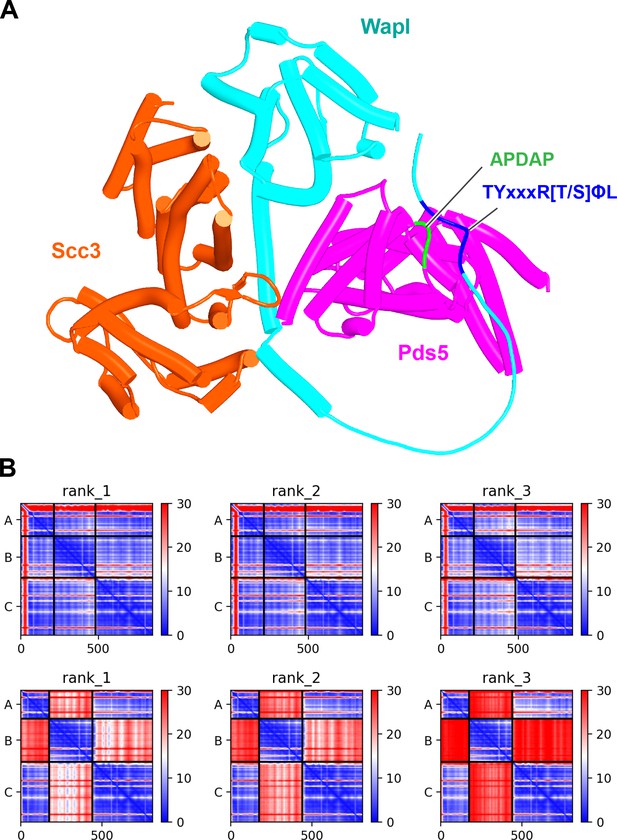
Formation of a canonical Pds5:Scc3:Wapl ternary complex in yeast.
(A) AlphaFold 2 (AF) prediction for a S. cerevisiae Pds5:Scc3:Wapl rigid body ternary complex accompanied by interaction between Wapl’s TYxxxR[T/S]ΦL motif and Pds5’s APDAP loop (f32B). (B) PAEs for the top three ranked models for S. cerevisiae’s Pds5:Scc3:Wapl ternary complex with (top row, f32B) and without (bottom row, f32A) Wapl’s TYxxxR[T/S]ΦL motif. Note that the lower confidence complexes predicted by AF for those lacking Wapl’s TYxxxR[T/S]ΦL motif are not of the canonical variety. In both (A) and (B), chains A, B, and C correspond to Wapl, Pds5, and Scc3, respectively. Interaction between Wapl’s TYxxxR[T/S]ΦL motif and Pds5 is represented by the thin blue lines on the left and upper squares of the top three maps in (B). Chains in the PAEs from f32B are Wapl (A), Pds5 (B), and Scc3 (C).
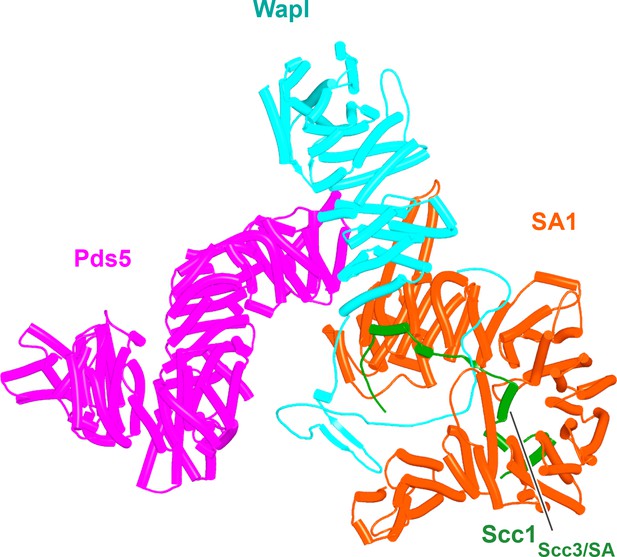
Composite model of the human Pds5:SA1:Wapl:Scc1 quarternary complex (f35) created using by superimposing rank 1 models from f23, f25, and f28.
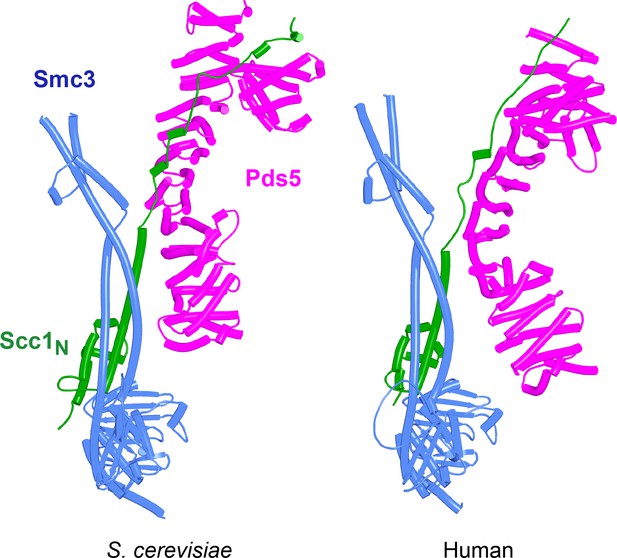
AlphaFold 2 (AF) predictions for the interaction of Scc1’s N-terminal 160 residues with Smc3 and with an N-terminal fragment of Pds5 for both S. cerevisiae (f36) and human (f37).
Chains in the PAEs from f36 are Smc3 (A), Smc3 (B), Scc1 (C), and Pds5 (D). Chains in the PAEs from f37 are Scc1 (A), Pds5 (B), Smc3 (C), and Smc3 (D).
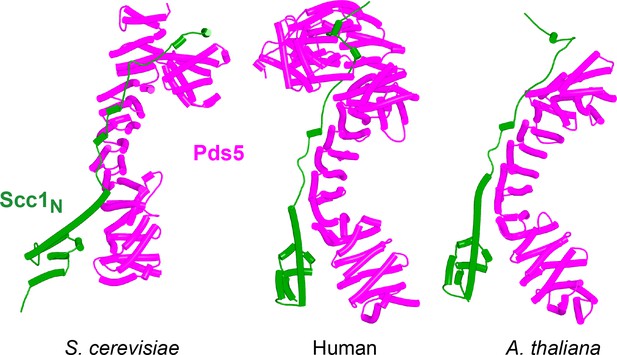
AlphaFold 2 (AF) predictions for the interaction between Scc1’s N-terminal 160 residues and Pds5 in S. cerevisiae (f38), humans (f39), and A. thaliana (f40).
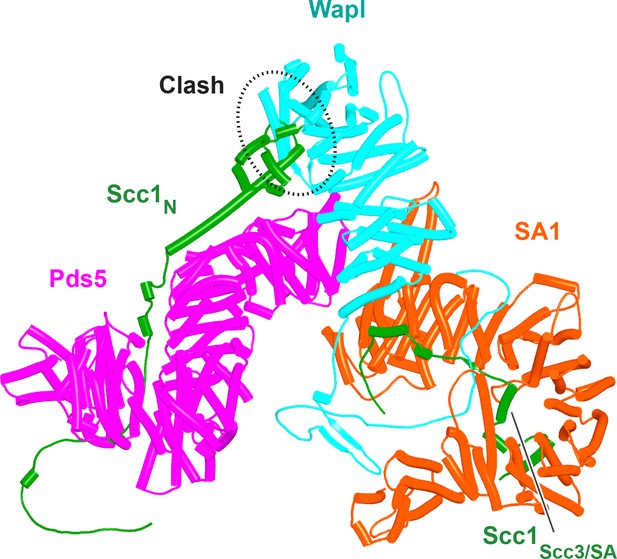
A putative quarternary complex between human Scc1N Pds5, SA1, and Wapl, created by super-imposing f23, f25, f28, and f39.
The Scc1’s NTD points toward, but clashes with the highly conserved cleft in Wapl’s CTD that is otherwise predicted to bind a motif PSCLSVCNVT (conserved among metazoa) located in Wapl’s N-terminal sequences (see Figure 26B).
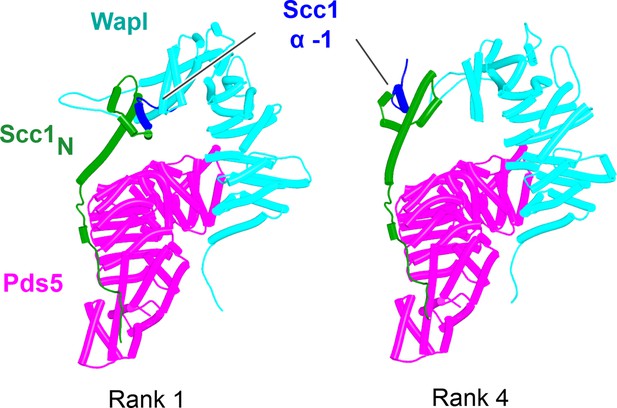
AlphaFold 2 (AF) predictions for ternary complexes (models ranks 1 and 4) containing Scc1N, Pds5’s NTD, and Wapl’s CTD (f42).
Chains in the PAEs from f42 are Scc1 (A), Pds5 (B), and Wapl (C).
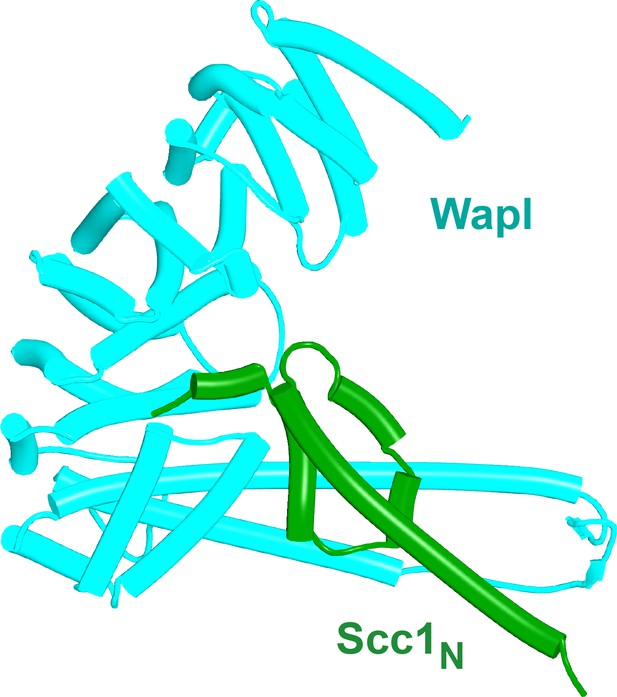
Interaction between Scc1's NTD and Wapl's CTD.
AlphaFold‘s prediction for the interaction of human Scc1’s NTD with Wapl’s CTD (f34A).
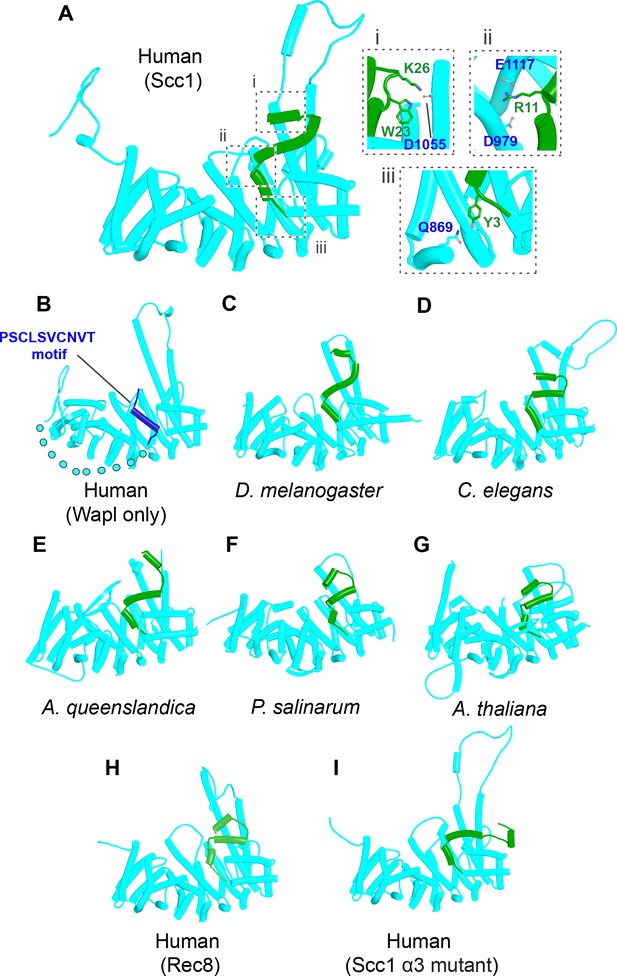
AlphaFold 2 (AF) predictions for the interactions between Wapl’s CTD and the N-terminal 35 amino acids of Scc1 for humans.
(A, f43), D. melanogaster (C, f44), C. elegans (D, f45), the sponge A. queenslandica (E, f46), the green alga P. salinarum (F, f47), and A. thaliana (G, f48). (B), AF prediction for human Wapl alone (AlphaFold Protein Structure Database AF-Q7Z5K2-F1). (H) AF prediction for the interaction between Wapl’s CTD and the N-terminal 36 amino acids of human Rec8 (f49). (I) Interaction between Wapl’s CTD and the N-terminal 35 amino acids of Scc1 in a ternary complex containing Scc1N(H58A, L59S, G62D, R65A, R69A), Pds5’s NTD, and Wapl’s CTD (f52A). See Figure 28 for the same structure in the complete ternary complex.
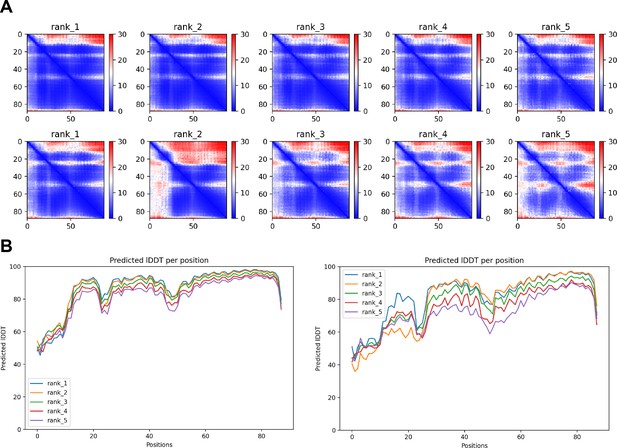
Mutations that weaken interaction between Scc1's first two alpha helices with its alpha 3 helix.
(A) PAEs for WT Scc1N (top, f50) and mutant Scc1N(H58A, L59S, G62D, R65A, R69A) (bottom, f51). (B) pLDDT plots for WT Scc1N (left, f50) and mutant Scc1N(H58A, L59S, G62D, R65A, R69A) (right, f51).
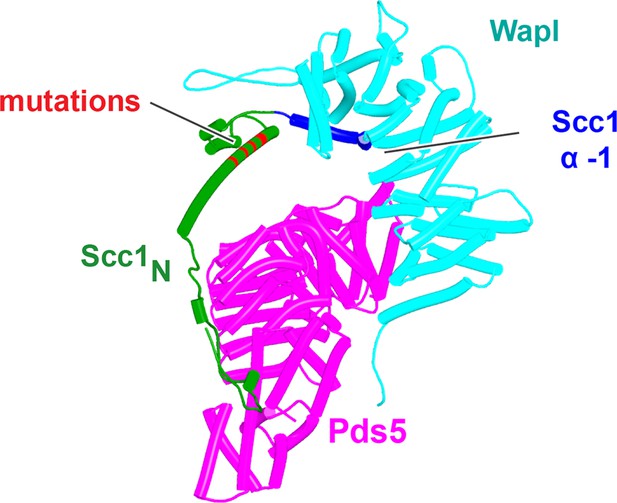
AlphaFold 2 (AF) prediction of a ternary complex containing Scc1N(H58A, L59S, G62D, R65A, R69A), Pds5’s NTD, and Wapl’s CTD (f52A).
The location of α3 Scc1N mutations that distinguish it from f42 shown in Figure 24 is marked in red. Chains in the PAEs from f52A are Scc1 (A), Pds5 (B), and Wapl (C).
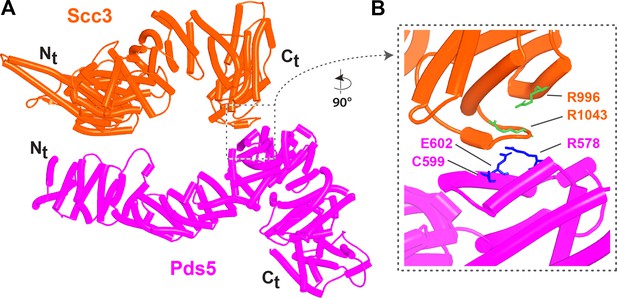
Interaction between Scc3 and Pds5 in yeast.
(A) AlphaFold 2 (AF) prediction for the interaction between Scc3 and Pds5 in S. cerevisiae (f54A). (B) Closeup showing residues whose mutation abrogates releasing activity (RA) and suppresses eco1Δ. Chains in the PAEs from f54A are Scc3 (A) and Pds5 (B).
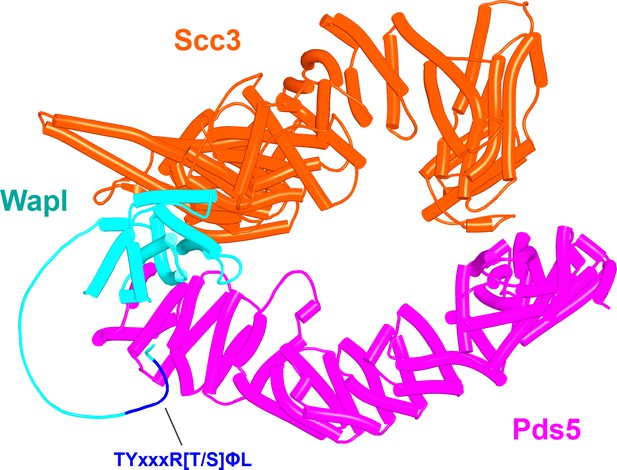
AlphaFold 2 (AF) prediction for a complex between S. cerevisiae Scc3, Pds5, and Wapl’s CTD with its associated TYxxxR[T/S]ΦL motif (f54B).
Chains in the PAEs from f54B are Wapl (A), Pds5 (B), and Scc3 (C).
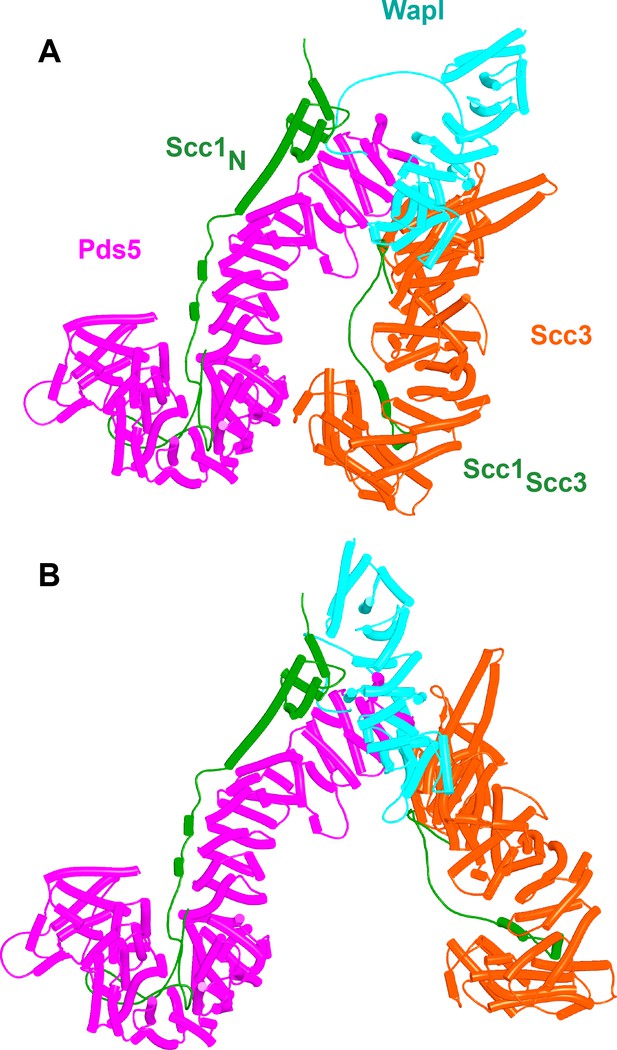
Putative quarternary complexes between yeast Scc1N, Pds5, Scc3, and Wapl.
(A) When formed with Scc3C interacting with Pds5P (f55), created by superimposing f24 and f38B onto f54B. (B) When formed without any interaction between Scc3C and Pds5P (f53), which involves a canonical rigid body Pds5:Scc3:Wapl ternary complex. Model created by super-imposing f24 and f38B onto f32B. A movie showing the transition between (A and B) at https://doi.org/10.6084/m9.figshare.22567525.v1.
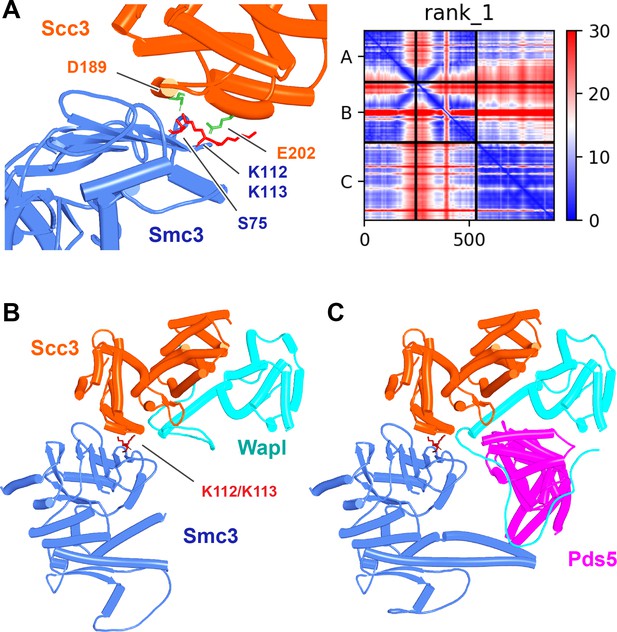
Interaction between Smc3’s ATPase and Scc3’s N-terminal domain.
(A) Left: detail of the interaction between Smc3 and Scc3 (f56A) showing Scc3(D189) and Scc3(E202) (marked green) and Smc3(S75, K112, K113) (marked red). (A) Right: PAEs for rank 1 model in A. Chains A and B correspond to Smc3 while C to Scc3. (B) Interaction between Smc3’s ATPase, Scc3’s N-terminal domain, and Wapl’s CTD (f56B). Chains in the PAEs from f56B are Smc3 (A), Smc3 (B), Scc3 (C), and Wapl (D). (C) Interaction of the canonical rigid body Pds5:Scc3:Wapl ternary complex with Smc3’s ATPase domain (f56C). Chains in the PAEs from f56C are Wapl (A), Pds5 (B), Scc3 (C), Smc3 (D), and Smc3 (E).
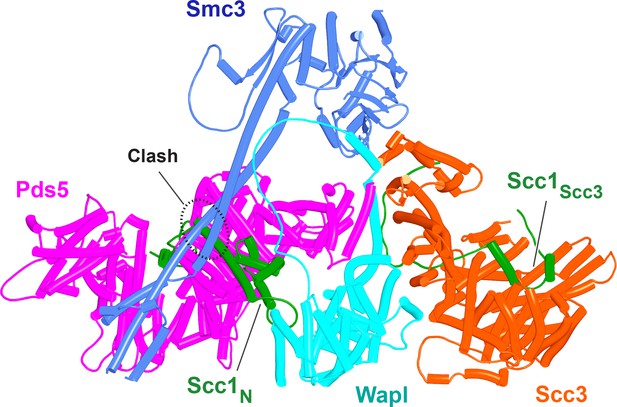
A model for the yeast quarternary Pds5:Scc3:Wapl:Scc1 complex in the canonical configuration when its Scc3 moiety is bound to the Smc3 ATPase head as predicted by AlphaFold 2 (AF), created by super-imposing f56A with the model in Figure 31B.
There is a modest clash between Scc1 and the Smc3 coiled coil that would presumably have been avoided if AF were able to predict the entire complex, which is beyond its capability.
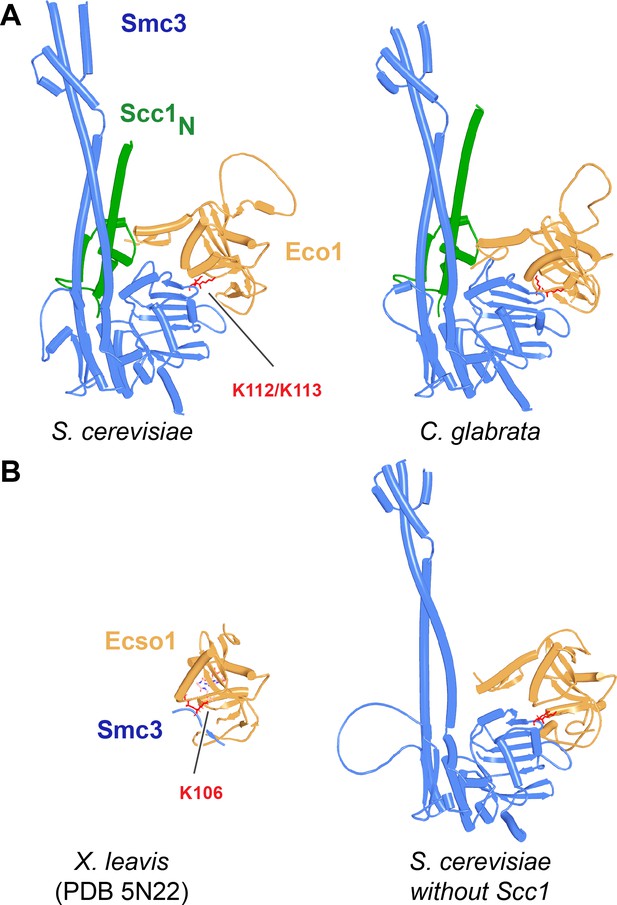
Interaction between Eco1/Esco1 and Smc3.
(A) AlphaFold 2 (AF) predictions for Smc3’s interaction with Eco1 from S. cerevisiae (f67) and C. glabrata (f68) with the Scc1 NTD present. (B) Left: similarity of this interaction with a crystal structure (PDB: 5N22) of Esco1 with a K106-CoA conjugate. A superimposition of f67 and 5N22 is shown in f69. Right: as in (A), left, without Scc1 NTD. Chains in the PAEs from f67A are Eco1 (A), Scc1 (B), Smc3 (C), and Smc3 (D).
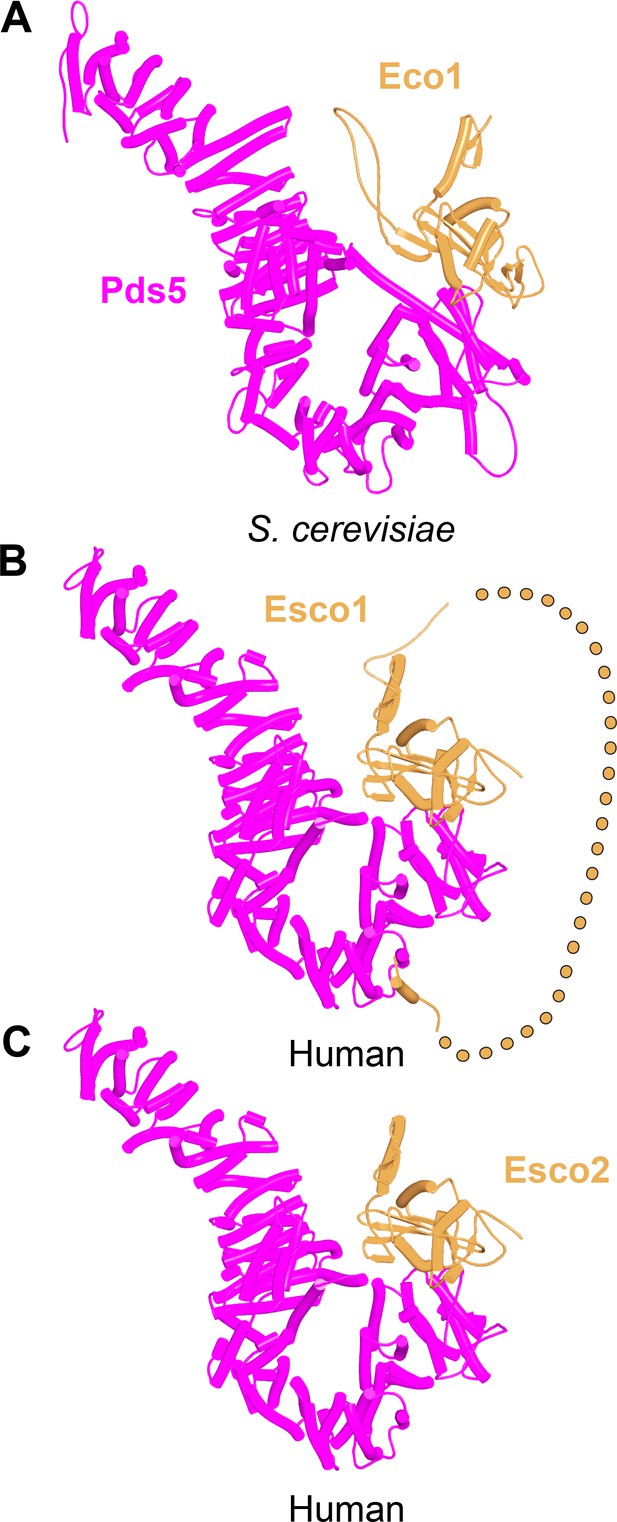
AlphaFold 2 (AF) predictions for interactions between Pds5 and S. cerevisiae Eco1 (A, f75), human Esco1 (B, f22), and human Esco2 (C, f76).
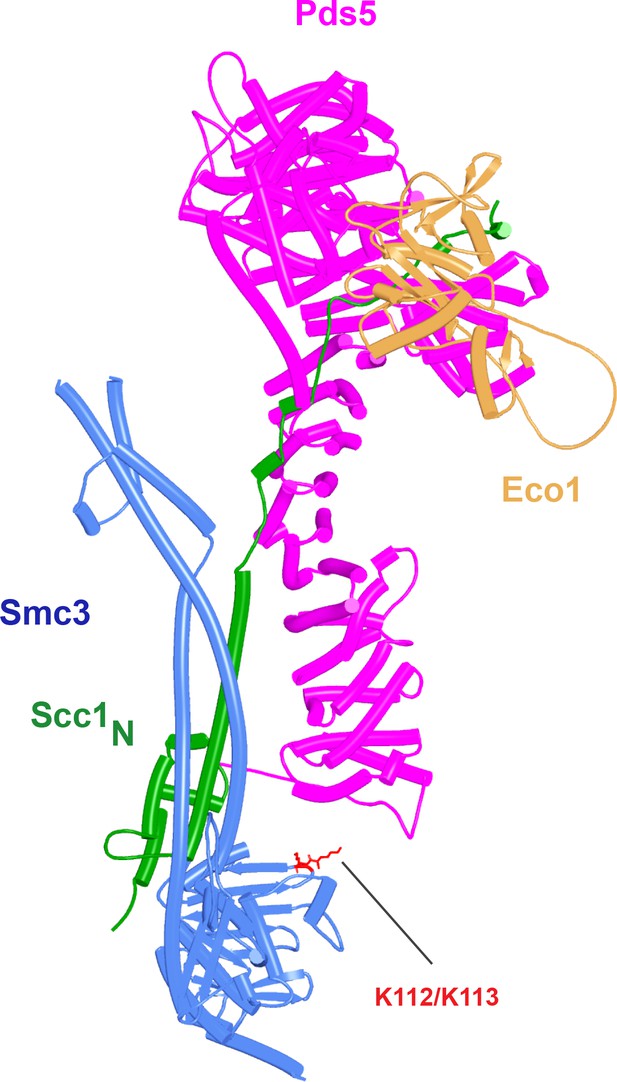
Projected position of Eco1 bound to Pds5 if Scc1N bound to the latter adopts the orientation predicted by AlphaFold 2 (AF) (f36).
The model was created by superimposing f36 and f75 via their Pds5 moieties. For Eco1 to bind Smc3 K112 and K113, while still bound to Pds5, the Scc1 sequences between Scc1N α3 and the part bound to Pds5 must bend by 180°, or Scc1’s NTD must dissociate from Smc3.
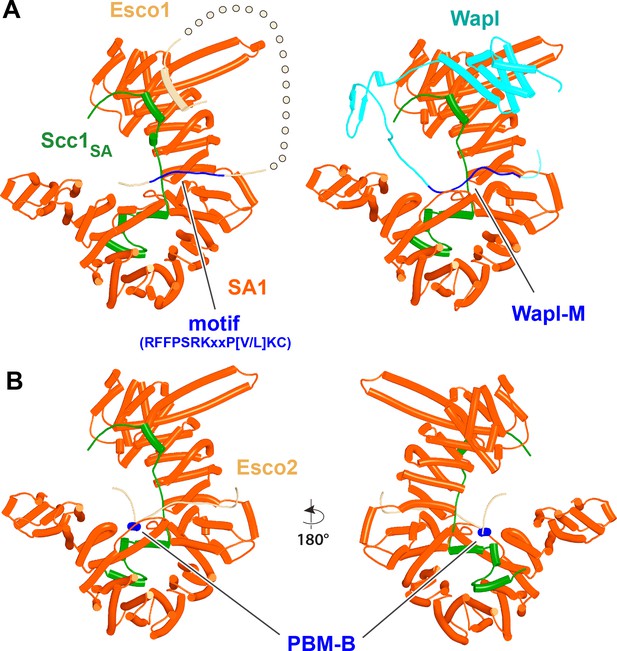
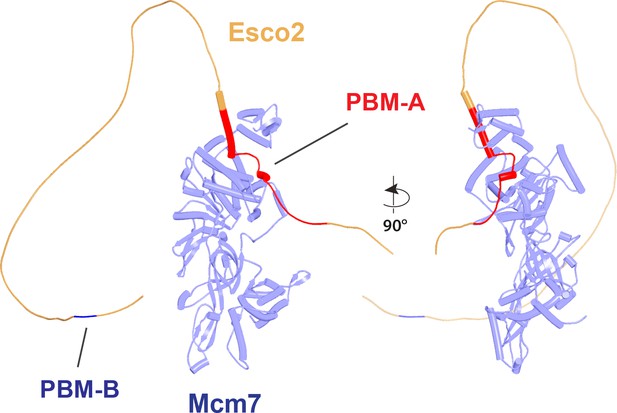
Interaction of Esco1 and Esco1 with SA1.
(A) AlphaFold 2 (AF) predictions for the interactions between SA1:Scc1 and the N-terminal extension of Esco1 (left, f77) and between SA2:Scc1 and Wapl-M (right, f25). (B) AF prediction for the interaction between SA2:Scc1 and Esco2’s PBM-B (f78). Chains in the PAEs from f77 are Scc1 (A), SA (B), and Esco1 (C). Chains in the PAEs from f78 are Esco2 (A), Scc1 (B), and SA (C).
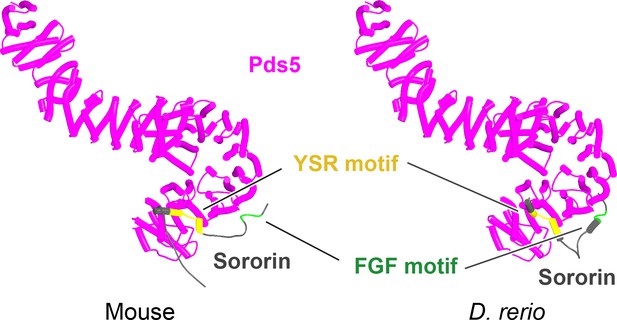
AlphaFold 2 (AF) predictions for Sororin’s interaction with Pds5B in mouse (f82) and zebrafish (f21).
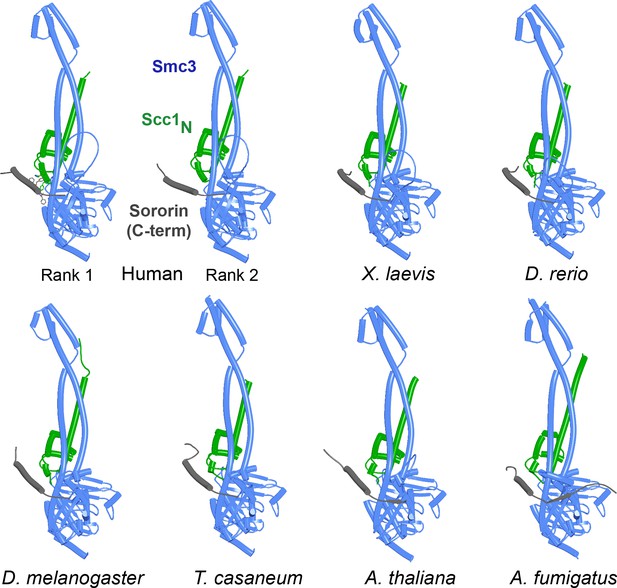
AlphaFold 2 (AF) predictions for Sororin’s interaction with Smc3 bound to Scc1’s NTD in humans (f84), X. laevis (f85), D. rerio (f86), D. melanogaster (f87), T. casaneum (f88), A. thaliana (89), and A. fumigatus (f90).
Chains in the PAEs from f84 are Sororin (A), Smc3 (B), Smc3 (C), and Scc1 (D).
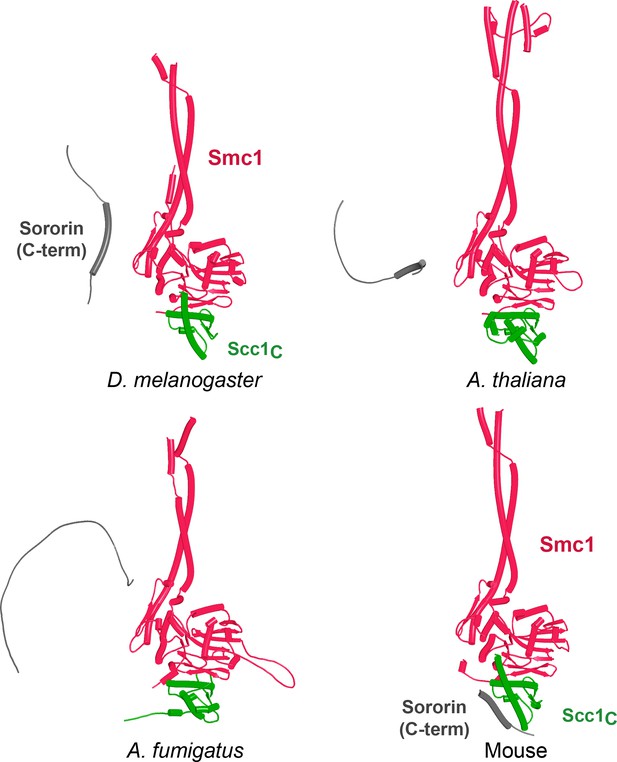
AlphaFold 2 (AF) predictions for the Sororin box’s interaction with a complex between Smc1 and Scc1’s CTD in D. melanogaster (f91), A. thaliana (f92), A. fumigatus (f93), and mouse (f94).