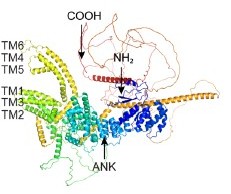
miR-252 targeting temperature receptor CcTRPM to mediate the transition from summer-form to winter-form of Cacopsylla chinensis
Figures
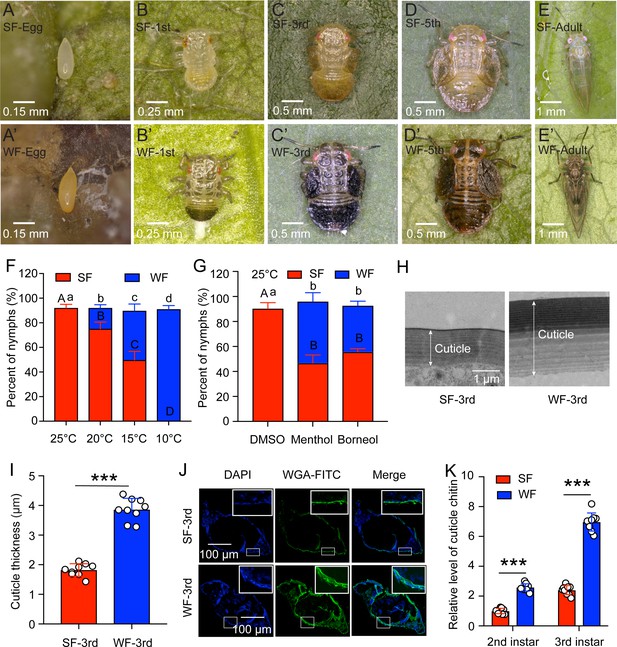
Low temperature induced the transition from summer form (SF) to winter form (WF) in C. chinensis.
(A-E and A’-E’) The morphology of egg, 1st, 3rd, and 5th instar nymph, 5 days adult for SF and WF of C. chinensis, respectively. The scale bars were labeled in the lower left of each figure. (F) Percent of SF and WF under different temperatures (25 °C, 20 °C, 15 °C, and 10 °C) when induction of newly-hatched (within 12 hr) 1st instar nymph of C. chinensis. (G) Effect of two chemical cooling agents (menthol and borneol) on the percent of SF and WF when induction of newly-hatched (within 12 hr) 1st instar nymph of C. chinensis under 25 °C. (H) Ultrastructure comparison of the nymph cuticle for SF 3rd instar and WF 3rd instar of C. chinensis using transmission electron microscopy. Scale bar is 1 μm. The two-way arrow indicated the cuticle thickness. (I) Comparison of the nymph cuticle thickness for SF 3rd instar and WF 3rd instar of C. chinensis. (J) Chitin staining of nymph cuticle with WGA-FITC in SF 3rd instar and WF 3rd instar of C. chinensis. DAPI: the cell nuclei were stained with DAPI and visualized in blue. WAG-FITC: the cuticle chitin was labeled with FITC and visualized in green. Merge: merged imaging of DAPI, and WAG-FITC signals. Scale bar is 100 μm. (K) Determination of cuticle chitin content in 2nd and 3rd instar nymph of SF and WF. The data of nymph percent were shown as the mean ± SD with three independent biological replications of at least 30 nymphs for each biological replication. Different letters above the bars indicated statistically significant differences (P<0.05), as determined by ANOVA followed with a Turkey’s HSD multiple comparison test in SPSS 20.0 software. The results of cuticle thickness and cuticle chitin content were presented as mean ± SD of three independent biological samples with three technical replications for each biological replication. Statistically significant differences were determined with the pair-wise Student’s t-test, and significance levels were denoted by *** (p<0.001).
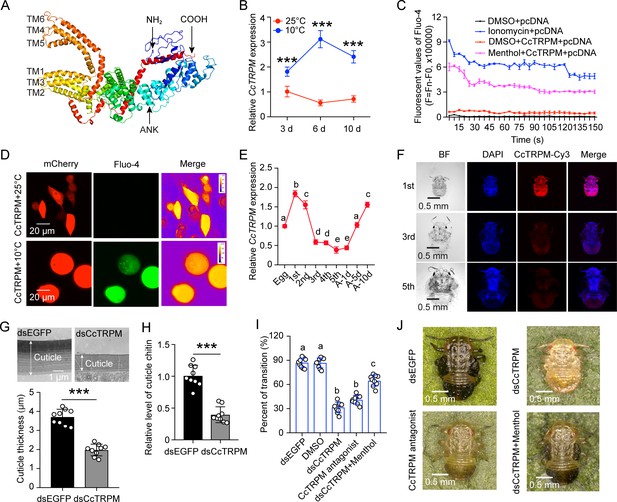
Temperature receptor CcTRPM modulated the transition from SF to WF of C. chinensis in response to low temperature.
(A) The predicted protein tertiary structure of CcTRPM. The conserved ankyrin repeat (ANK) domain was indicated in the N-terminal. The conserved six transmembrane domain of ion channels structure were shown as TM1-TM6. (B) The mRNA expression of CcTRPM in response to different temperatures of 25°C and 10°C by qRT-PCR. (C) Fluorescence detection of Fluo-4 AM after heterologous expression of CcTRPM in mammalian HEK293T cells in response to menthol treatment. The recombinant plasmid was generated by inserting the full ORF sequence of CcTRPM into pcDNA3.1(+)-mCherry plasmid. DMSO treatment and ionomycin treatment were used as negative control and positive control, separately. (D) Representative images of Ca2+ imaging after heterologous expression of CcTRPM in mammalian HEK293T cells in response to different temperature treatment. “CcTRPM +10 °C” means the recombinant plasmid of CcTRPM with pcDNA3.1(+)-mCherry was treated with 10 °C. “CcTRPM +25 °C” denotes the pcDNA3.1(+)-mCherry plasmid with CcTRPM was treated with 25 °C. mCherry and Fluo-4 signal are shown in red and green, respectively. Scale bar is 20 μm. (E) The developmental expression pattern of CcTRPM for SF at mRNA level using qRT-PCR. 1st, 2nd, 3rd, 4th, and 5th are the nymphs at the first, second, third, fourth and fifth instar, respectively. A-1d, A-5d, and A-10d are the adults at 1 day, 5 days, and 10 days, separately. (F) Representative confocal images of CcTRPM in different developmental stages of SF using FISH. Scale bar is 0.5 mm. BF: the bright field. DAPI: the cell nuclei were stained with DAPI and visualized in blue. CcTRPM-Cy3: CcTRPM signal was labeled with Cy3 and visualized in red. Merge: merged imaging of BF, DAPI, and CcTRPM-Cy3 signals. (G–H) Comparison of the nymph cuticle ultrastructure, cuticle thickness, and cuticle chitin content of SF 1st instar treated with dsCcTRPM and dsEGFP at 15 days. (I) The transition percent of SF 1st instar nymphs treated with dsEGFP, DMSO, dsCcTRPM, CcTRPM antagonist and dsCcTRPM +menthol at 15 days under 10 °C. For RNAi experiments, summer-form 1st instar nymphs were fed with dsEGFP (500 ng/μL) or dsCcTRPM (500 ng/μL). To mimic RNAi effect, summer-form 1st instar nymphs were fed with 0.1% DMSO or CcTRPM antagonist (20 ng/μL). For the rescue experiment, summer-form 1st instar nymphs were fed with the mixture of dsCcTRPM (500 ng/μL) and menthol (1 mg/mL). Then, counted the number of summer-form and winter-form individuals and calculated the transition percent. (J) The phenotypes of SF 1st instar nymphs treated with dsEGFP, dsCcTRPM, CcTRPM antagonist and dsCcTRPM +menthol at 15 days under 10 °C. The data in 2B and 2E are shown as the mean ± SD with three independent biological replications of at least 30 nymphs for each biological replication. Scale bar is 0.5 mm. Data in 2 G and 2 H are presented as mean ± SD with three biological replications of three technical replicates for each biological replication. Data in 2I are presented as mean ± SD with nine biological replications. Statistically significant differences were determined with the pair-wise Student’s t-test, and significance levels were denoted by *** (p<0.001). Different letters above the bars indicated statistically significant differences (p<0.05), as determined by ANOVA followed with a Turkey’s HSD multiple comparison test in SPSS 20.0 software.
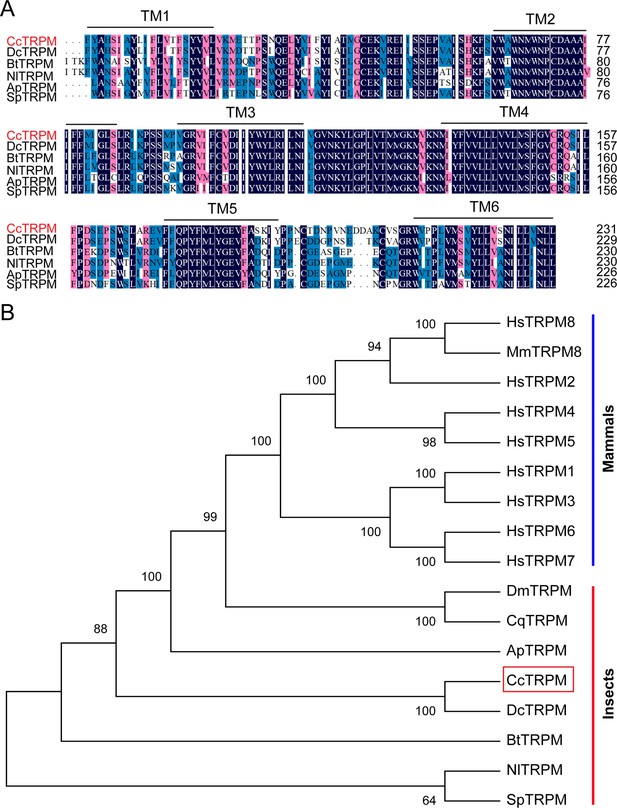
Sequence characterization of TRPM from different insect species.
(A) Multiple alignments of the amino acid sequences of CcTRPM transmembrane domain with homologs from other insect species. Black represents 100% identity, red represents 75% identity and blue represents <75% identity. The transmembrane domain of TM1 to TM6 are indicated by black horizontal line. CcTRPM (C. chinensis, OQ658558), DcTRPM (Diaphorina citri, XP_017299512.2), BtTRPM (Bemisia tabaci, XP_018904971.1), NlTRPM (Nilaparvata lugens, AOR81476.1), ApTRPM (Acyrthosiphon pisum, XP_003245193.1), SpTRPM (Schistocerca piceifrons, XP_047116036.1). The same as below. (B) Phylogenetic tree analysis of CcTRPM and its homologs in other insect species. HsTRPM1 (Homo sapiens, NP_001238949.1), HsTRPM2 (Homo sapiens, NP_001307279.2), HsTRPM3 (Homo sapiens, NP_001353070.1), HsTRPM4 (Homo sapiens, NP_060106.2), HsTRPM5 (Homo sapiens, NP_055370.1), HsTRPM6 (Homo sapiens, NP_060132.3), HsTRPM7 (Homo sapiens, NP_060142.3), HsTRPM8 (Homo sapiens, NP_001384540.1), MmTRPM8 (Mus musculus, AAL79553.1), DmTRPM (Drosophila melanogaster, NP_001401007.1), CqTRPM (Culex quinquefasciatus, XP_038115236.1).
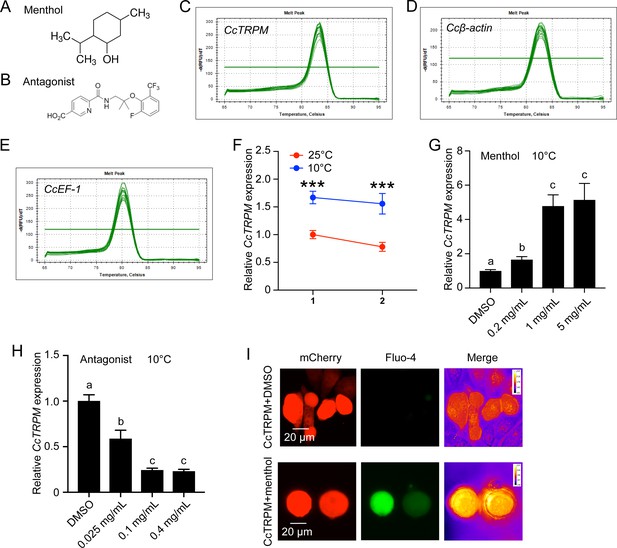
Effect of menthol and CcTRPM antagonist treatment on the mRNA expression of CcTRPM under 10 °C condition.
(A–B) The chemical structure of menthol and TRPM antagonist. (C–E) Melting curve for qRT-PCR primers of CcTRPM, Ccβ-actin and CcEF-1. (F) The mRNA expression of CcTRPM in response to different temperatures of 25°C and 10°C at 1 day and 2 days by qRT-PCR. (G) Effect of menthol treatment at different concentrations on the mRNA expression of CcTRPM under 10 °C condition. (H) Effect of TRPM antagonist treatment at different concentrations on the mRNA expression of CcTRPM under 10 °C condition. (I) Representative images of Ca2+ imaging after heterologous expression of CcTRPM in mammalian HEK293T cells in response to menthol treatment. ‘CcTRPM +menthol’ means the recombinant plasmid of CcTRPM with pcDNA3.1(+)-mCherry was treated with menthol. “CcTRPM +DMSO” denotes the pcDNA3.1(+)-mCherry plasmid with CcTRPM was treated with DMSO. mCherry and Fluo-4 signal are shown in red and green, respectively. Scale bar is 20 μm. Data are shown as mean ± SD with three biological replications. Statistically significant differences were determined with the pair-wise Student’s t-test, and significance levels were denoted by *** (p<0.001). Different letters above the bars indicated statistically significant differences (p<0.05), as determined by ANOVA followed with a Turkey’s HSD multiple comparison test in SPSS 20.0 software.
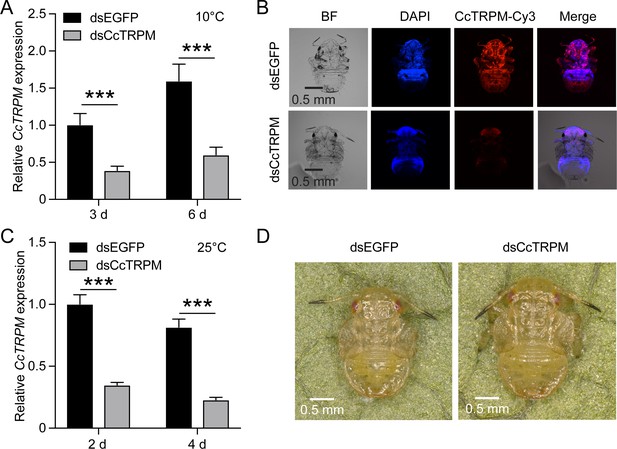
RNAi efficiency of CcTRPM and effect of CcTRPM knockdown on the phenotype at 25 °C.
(A) RNAi efficiency of CcTRPM after dsCcTRPM treatment at 3 days and 6 days by qRT-PCR under 10 °C condition. (B) Representative confocal images of CcTRPM after SF 1st instar nymphs treated with dsCcTRPM for 6 days at 10 °C by FISH. Scale bar is 0.5 mm. BF: the bright field. DAPI: the cell nuclei were stained with DAPI and visualized in blue. CcTRPM-Cy3: CcTRPM signal was labeled with Cy3 and visualized in red. Merge: merged imaging of BF, DAPI, and CcTRPM-Cy3 signals. (C) RNAi efficiency of CcTRPM after dsCcTRPM treatment at 2 days and 4 days by qRT-PCR under 25 °C condition. (D) The phenotype of SF 1st instar nymphs treated with dsEGFP and dsCcTRPM at 10 days under 25 °C condition. Scale bar is 0.5 mm. Data are shown as mean ± SD with three biological replications. Statistically significant differences were determined with the pair-wise Student’s t-test, and significance levels were denoted by *** (p<0.001).
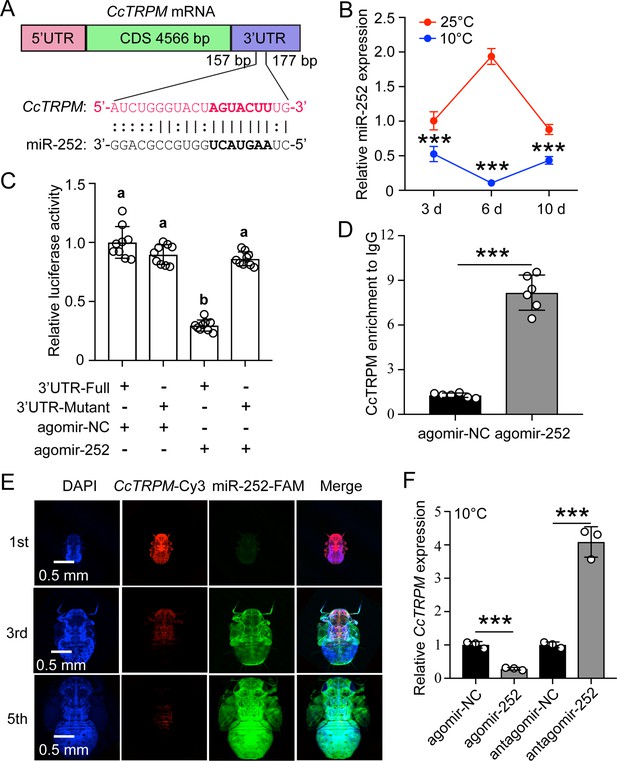
Confirming of miR-252 targeted CcTRPM.
(A) The putative miR-252 binding sites in CcTRPM 3’UTR were predicted by miRanda and Targetscan. (B) The expression profiles of miR-252 in response to different temperatures of 25°C and 10°C by qRT-PCR. (C) In vitro validation of the target interactions between miR-252 and CcTRPM by dual luciferase reporter assays. (D) In vivo demonstration of miR-252 targeting CcTRPM by RNA-binding protein immunoprecipitation (RIP) assay. (E) Representative confocal images of miR-252 and CcTRPM in 1st, 3rd and 5th instar of SF. Scale bar is 0.5 mm. The signals of DAPI and CcTRPM-Cy3 are same as the above describing. miR-252-FAM: the miR-252 signal was labeled with FAM and visualized in green. Merge: merged imaging of co-localization of cell nucleus, CcTRPM and miR-252. (F) Effect of miR-252 agomir and antagomir treatment 3 days on the expression of CcTRPM at mRNA level. The data in 3B and 3 F are shown as the mean ± SD with three independent biological replications of at least 30 nymphs for each biological replication. Data in 3 C are presented as mean ± SD with nine biological replications. Data in 3D are presented as mean ± SD with two biological replications of three technical replicates for each biological replication. Statistically significant differences were determined with the pair-wise Student’s t-test, and significance levels were denoted by *** (p<0.001). Different letters above the bars indicated statistically significant differences (p<0.05), as determined by ANOVA followed with a Turkey’s HSD multiple comparison test in SPSS 20.0 software.
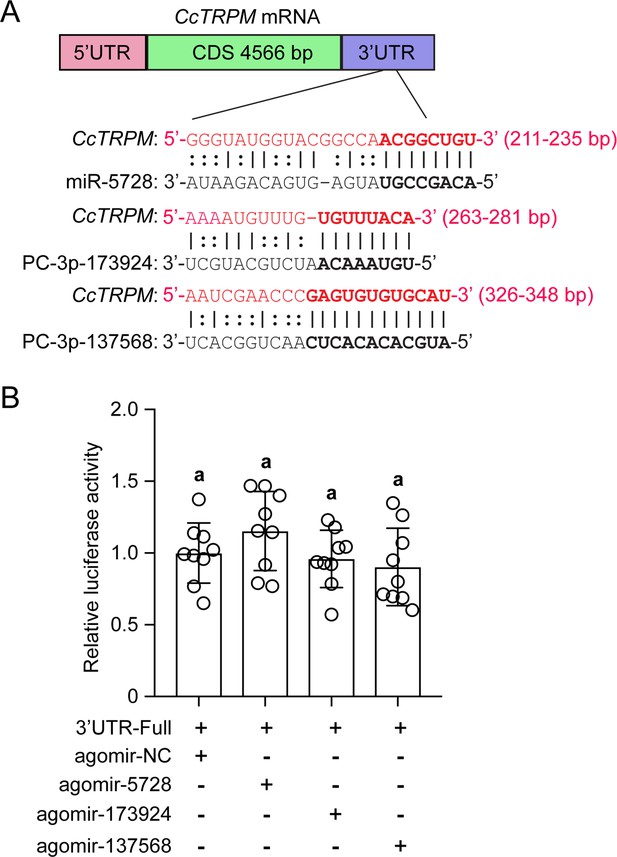
Validation of the target interactions between CcTRPM and other three miRNAs.
(A) The putative binding sites of miR-5728, PC-3p-173924, and PC-3p-137568 in CcTRPM 3’UTR were predicted by miRanda and Targetscan. (B) In vitro validation of the target interactions between miR-5728, PC-3p-173924, and PC-3p-137568 with CcTRPM by dual luciferase reporter assays. Data are presented as mean ± SD with nine biological replications. Different letters above the bars indicated statistically significant differences (p<0.05), as determined by ANOVA followed with a Turkey’s HSD multiple comparison test in SPSS 20.0 software.
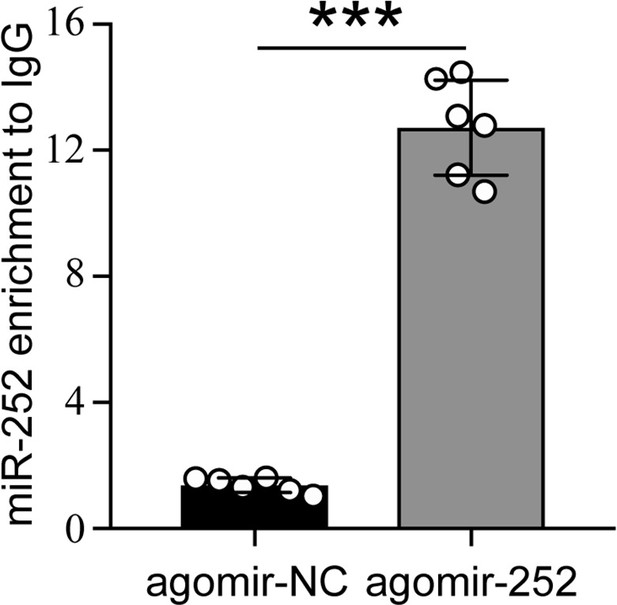
Transcript of miR-252 was significantly enriched by antibody against Ago1 in agomir-252 treated group compared with agomir-NC group.
Data are shown as mean ± SD with two biological replications of three technical replicates for each biological replication. Statistically significant differences were determined with the pair-wise Student’s t-test, and significance levels were denoted by *** (p<0.001).
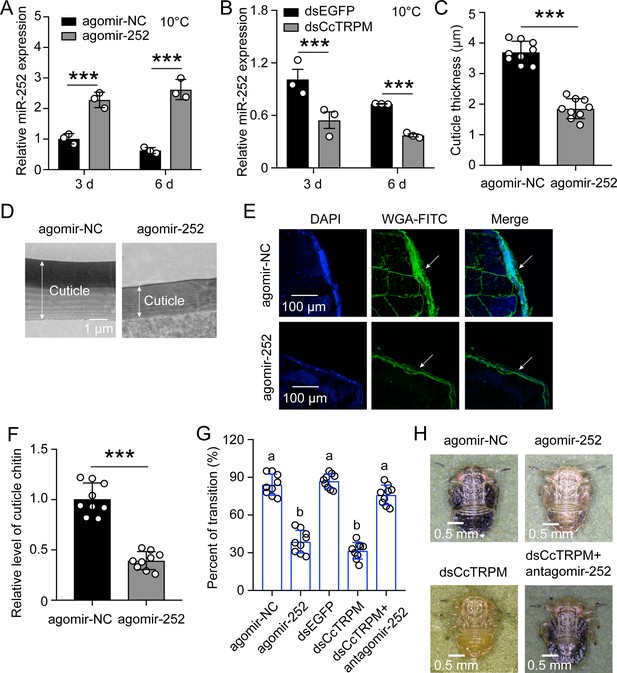
miR-252 mediated the transition from SF to WF in C. chinensis by suppressing CcTRPM.
(A) Effect of miR-252 agomir treatment on the mRNA expression of miR-252 after 3 days and 6 days under 10 °C. agomir-NC treatment was used as the control. (B) The mRNA expression of miR-252 after dsCcTRPM treatment 3 days and 6 days compare with the dsEGFP treatment under 10 °C. (C–F) Comparison of the nymph cuticle thickness, cuticle ultrastructure, cuticle chitin staining with WGA-FITC, and chitin content of SF 1st instar after treatment with agomir-NC and agomir-252 at 15 days. Scale bar in (D) is 1μm and in (E) is 100 μm. The two-way arrow indicated the cuticle thickness. The DAPI and WAG-FITC signals were same as the above describing. (G) The transition percent of SF 1st instar nymphs treated with agomir-NC, agomir-252, dsEGFP, dsCcTRPM, and dsCcTRPM +antagomir-252 at 15 day under 10 °C. (H) The phenotypes of SF 1st instar nymphs treated with agomir-NC, agomir-252, dsCcTRPM, and dsCcTRPM +antagomir-252 at 15 day under 10 °C. Scale bar is 0.5 mm. The data in 4A and 4B are shown as the mean ± SD with three independent biological replications of at least 30 nymphs for each biological replication. Data in 4C and 4F are presented as mean ± SD with three biological replications of three technical replications for each biological replication. Data in 4G are presented as mean ± SD with nine biological replications. Statistically significant differences were determined with the pair-wise Student’s t-test, and significance levels were denoted by *** (p<0.001). Different letters above the bars indicated statistically significant differences (p<0.05), as determined by ANOVA followed with a Turkey’s HSD multiple comparison test in SPSS 20.0 software.
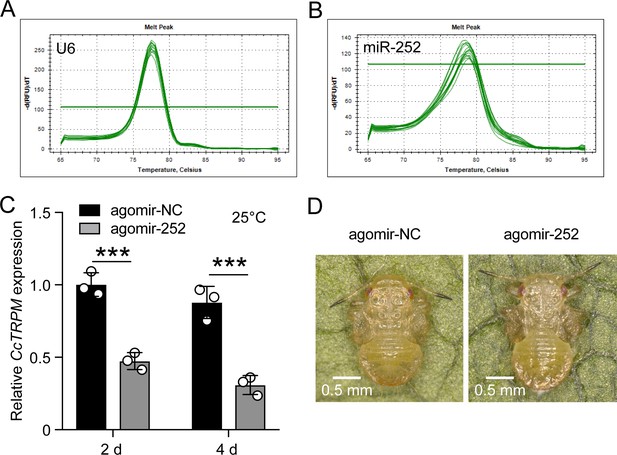
Effect of agomir-252 on the phenotype of SF 1st instar nymphs at 25 °C.
(A–B) Melting curve for qRT-PCR primers of U6 and miR-252. (C) Effect of agomir-252 treatment 2 day and 4 day on the mRNA expression of CcTRPM under 25 °C condition. (D) The phenotype of SF 1st instar nymphs treated with agomir-NC and agomir-252 at 10 day at 25 °C. Scale bar is 0.5 mm. Data are shown as the mean ± SD with three independent biological replications of at least 30 nymphs for each biological replication. Statistically significant differences were determined with the pair-wise Student’s t-test, and significance levels were denoted by *** (p<0.001).
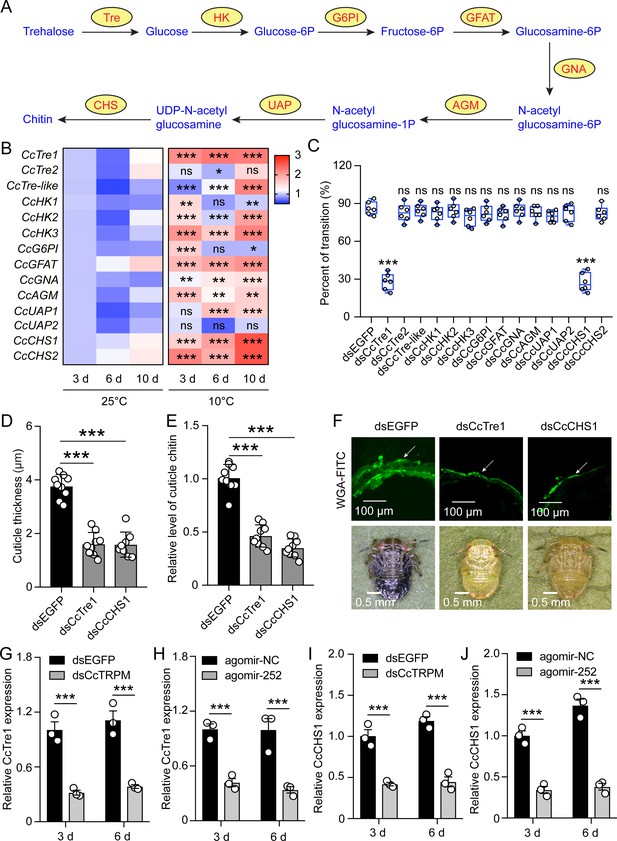
Chitin biosynthesis signaling involved in the transition from SF to WF of C. chinensis in response to low temperature and CcTRPM.
(A) Diagram of de novo biosynthesis of insect chitin. Tre: trehalase. HK: hexokinase. G6PI: glucose-6-phosphate isomerase. GFAT: fructose-6-phosphate aminotransferase. GNA: glucosamine-6-phosphate acetyltransferase. AGM: N-acetylglucosamine phosphate mutase. UAP: UDP- N-acetylglucosamine pyrophosphorylase. CHS: chitin synthase. (B) A heat map was constructed from the expression levels of chitin biosynthesis enzyme transcripts in C. chinensis after 25°C and 10°C treatment at 3 d, 6 d, and 10 d. (C) Effect of RNAi knockdown of 14 transcripts covering all the enzymes in the chitin biosynthesis pathway on the transition percent of SF 1st instar nymphs under 10 °C. (D–F) Comparison of the nymph cuticle thickness, cuticle chitin content, and cuticle chitin staining with WGA-FITC of SF 1st instar after treatment with dsEGFP, dsCcTre1, and dsCcCHS1 at 15 d. The WAG-FITC signal was same as the above describing. Scale bar in (F) is 100 μm and 0.5 mm, respectively. (G–H) Effect of dsCcTRPM and agomir-252 treatments on the mRNA expression of CcTre1 at 3 day and 6 day under 10 °C. (I–J) Effect of dsCcTRPM and agomir-252 treatments on the mRNA expression of CcCHS1 at 3 day and 6 day under 10 °C. The data in (C) are shown as the mean ± SD with six independent biological replications of at least 30 nymphs for each biological replication. Data in (D and E) are presented as mean ± SD with three biological replications of three technical replications for each biological replication. Data in (G-J) are presented as mean ± SD with three biological replications. Statistically significant differences were determined with the pair-wise Student’s t-test, and significance levels were denoted by * (p<0.05), ** (p<0.01), and *** (p<0.001).
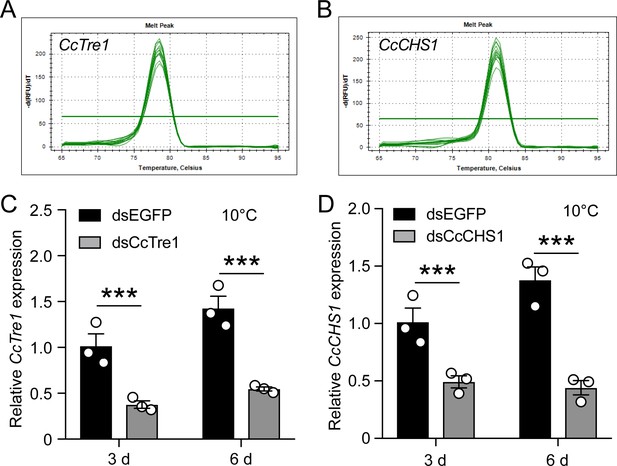
RNAi efficiency of CcTre1 and CcCHS1 at 10 °C by qRT-PCR.
(A–B) Melting curve for qRT-PCR primers of CcTre1 and CcCHS1. (C–D) RNAi efficiency of CcTre1 and CcCHS1 after dsCcTre1 and dsCcCHS1 treatment at 3 day and 6 day under 10 °C condition by qRT-PCR, respectively. Data are shown as mean ± SD with three biological replications. Statistically significant differences were determined with the pair-wise Student’s t-test, and significance levels were denoted by *** (p<0.001).
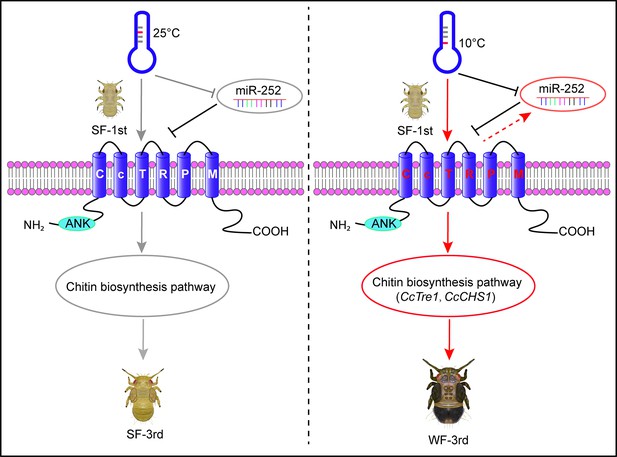
A model showing the key roles of miR-252 targeting CcTRPM to regulate the transition from SF to WF of C. chinensis in response to low temperature.
Under 25 °C condition, the temperature receptor CcTRPM was inactive as inhibiting by miR-252 in 1st instar nymphs of SF. The 1st instar nymphs of SF normally developed into 3rd instar nymphs of SF. However, under 10 °C condition, low temperature profoundly activated CcTRPM expression, miR-252 functioned as a negative regulator to mediate the expression of CcTRPM in case of over-expression in response to low temperature. Then, CcTRPM significantly increased the activity of chitin biosynthesis pathway, especially CcTre1 and CcCHS1, and led to cuticle chitin content was obvious raise and cuticle thickness became thicker. Finally, the 1st instar nymphs of SF developed to 3rd instar nymphs of WF in order to better adapt to low temperature. Red arrows and black T-bars indicated the activation and inhibition, respectively. Gray arrows and T-bars represented the inactive states of the genes or physiological processes, separately.
Additional files
-
Supplementary file 1
The primers used in current study and effect of miR-252 on the transition percent of SF 1st instar nymphs.
(a) The primers used in current study. (b) Effect of agomir-252 and antagomir-252 treatment on the transition percent of SF 1st instar nymphs under 10 °C.
- https://cdn.elifesciences.org/articles/88744/elife-88744-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/88744/elife-88744-mdarchecklist1-v1.pdf