The Toxoplasma monocarboxylate transporters are involved in the metabolism within the apicoplast and are linked to parasite survival
Figures
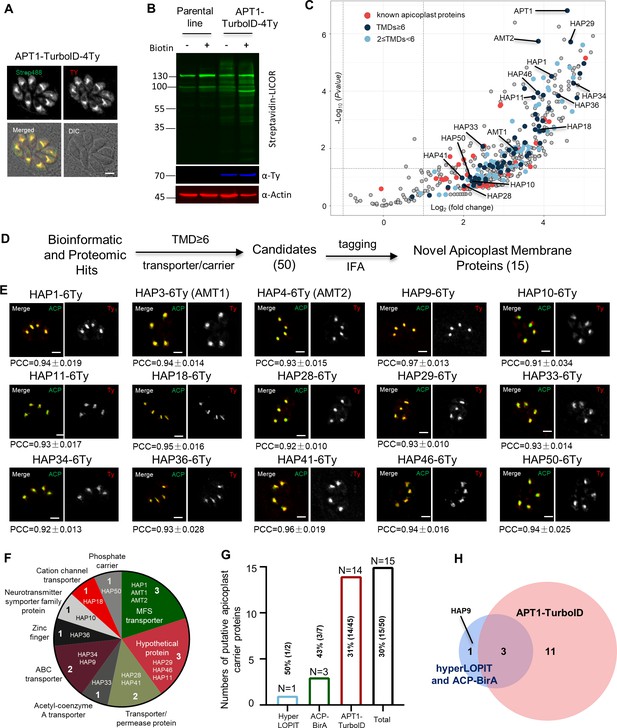
Discovery of apicoplast membrane proteins.
Activity test of biotinylation of proximal proteins by APT1-TurboID-4Ty. Parasites were grown in 500 μM D-biotin for 90 min, followed by detection of biotinylated proteins (green) by streptavidin reagents on indirect fluorescence assay (IFA) (A) and on western blots (B). Actin served as the loading control. Scale = 5 μm. (C) Volcano plot analysis comparing the TurboID fusion to the parental line. Three replicate mass-spectrometry experiments were analyzed by the Student t-test. Known apicoplast proteins (red) and candidates with different numbers of transmembrane domains (TMDs; deep and light blue were indicated, and novel apicoplast proteins were pointed out; see Supplementary file 2a). (D) Workflow of the discovery of novel apicoplast membrane proteins. Candidates from hyperLOPIT (Barylyuk et al., 2020), ACP-BirA (Boucher et al., 2018), and APT1-TurboID (this study) were screened by epitopte tagging and IFA. (E) Confocal co-localization of HAP-6Ty with the apicoplast marker ACP, showing confocal imaging of HAP-6Ty (red) and ACP (green), and Pearson correlation coefficiency (PCC). The PCC values over the merged fluoresent foci for candidate proteins and ACP were analyzed by a co-localization and region of interest (ROI) intensity analysis module in the NIS Elements AR software and shown with mean ± standard error of the mean (SEM; N = 6 for each). Scale = 5 μm. (F–H) Summary and comparison of the screening from three datasets. The putative transporters are grouped into diverse types (F) and identified from three different datasets (G), which shared three novel proteins, as shown by the Venn diagram (H). Three independent experiments were performed with similar outcomes (A, B, E).
-
Figure 1—source data 1
PDF file containing the uncropped western blot gels for verification of the TurboID fusion lines in Figure 1A.
- https://cdn.elifesciences.org/articles/88866/elife-88866-fig1-data1-v1.zip
-
Figure 1—source data 2
Excel file containing the raw images of confocal analysis for verification of co-localization of newly discovered proteins with the apicoplast marker ACP.
- https://cdn.elifesciences.org/articles/88866/elife-88866-fig1-data2-v1.zip
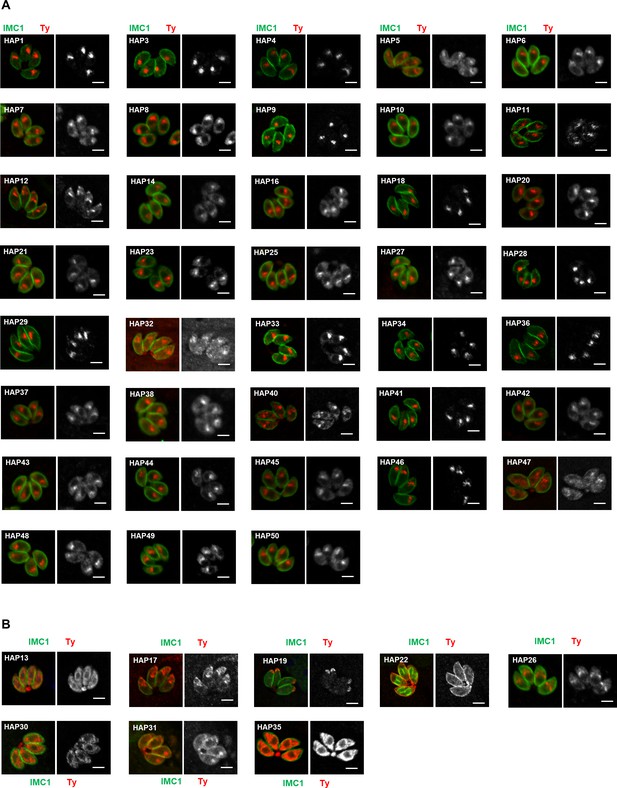
Indirect fluorescence assay (IFA) analysis of HAP-6Ty fusions in parasites.
(A, B) Candidates from hyperLOPIT, ACP-BirA, and APT1-TurboID were endogenously tagged with 6Ty at their C-termini using a CRISPR-Cas9 approach, followed by IFA analysis with antibodies against the inner membrane complex protein IMC1 (green) and Ty (red). Many of the HAPs were localized to a foci similar to position of the apicoplast in parasites (A), whereas others were localized to the cytosolic vesicles (e.g. HAP30, HAP13, and AP35), pellicular membrane (e.g. HAP22), or other structures (B). Scale = 5 μm. Note that HAP2, 15, 24, and 39 were not successfully tagged for IFA analysis. Three independent experiments were performed with similar outcomes and representative images are shown.
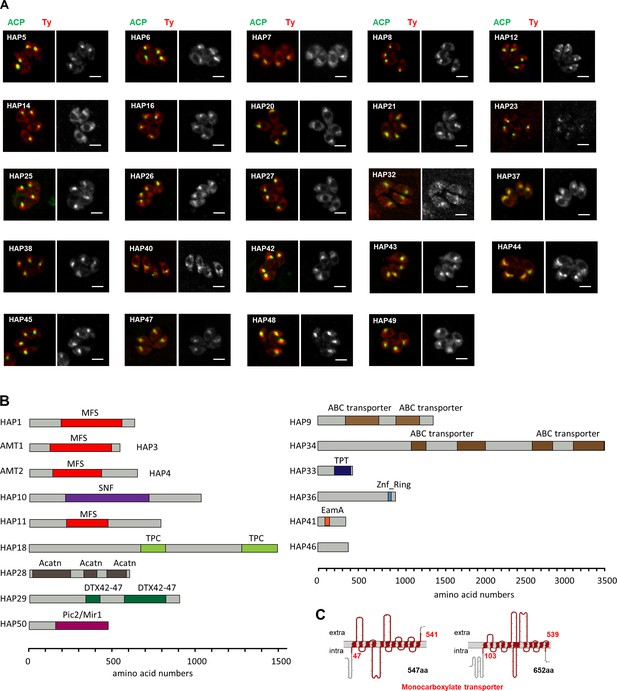
Analyses of non-apicoplast-localized HAP fusions, and domain analysis of novel apicoplast transporters.
(A) Shown here are the HAP-Ty fusions that were not localized to the apicoplast. Indirect fluorescence assay (IFA) analyses were performed with the HAP-6Ty fusions (red) and the apicoplast marker ACP (green). Scale = 5 μm. (B) Domain analysis of newly identified apicoplast membrane proteins. Proteins were analyzed by IntroProScan (https://www.ebi.ac.uk/interpro/about/interproscan/). MFS, major facillitator transporter superfamily capable of transporting small solutes in response to chemiosmotic ion gradients (Pao et al., 1998; Walmsley et al., 1998); SNF, sodium:neutrotransmitter symporter superfamily that is responsible for exitatory amino acid transport (Malandro and Kilberg, 1996); TPC, two-pore channels that have roles in organelle integrity, inter-organelle communication and growth in T. gondii (Li et al., 2021); acatn, acetyl-coenzyme A transporter (Bora et al., 1999); DTX42-47, among which DTX43 is a citrate transporter responsible for loading citrate into xylem tissues and facilitate iron transport to shoots (Green and Rogers, 2004; Rogers and Guerinot, 2002); ABC, ABC transporters that are involved in export or import of a wide variety of substrates ranging from small ions to macromolecules; Pic2/Mir1-like, phosphate carriers that were reported to import copper and inorganic phosphate into mitochondria (Hamel et al., 2004; Vest et al., 2013); TPT, Sugar phosphate transporter domain with a specificity for triose phosphate (Jack et al., 2001); Znf ring, Zinc finger that may bind metals, such as iron, or no metal at all; EamA, found in many members classed as drug/metabolite transporters (Jack et al., 2001). Protein lengths were scaled by amino acid numbers (aa). (C) The topology of two monocarboxylate transporters (AMT1 and AMT2) was analyzed by an online server PROTTER (https://wlab.ethz.ch/protter/start/).
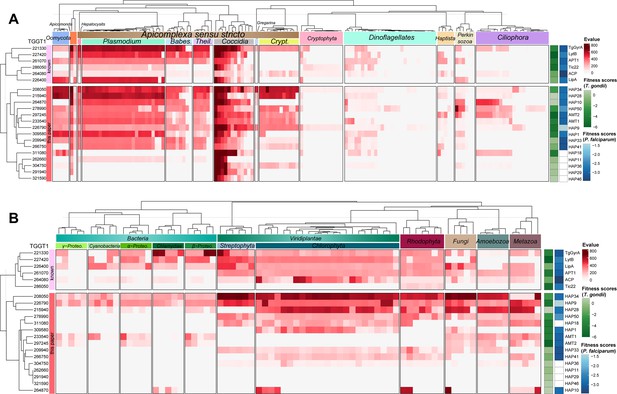
Conservation analysis of putative transporters.
Heatmap indicating conservation of known apicoplast proteins and newly identified proteins among chromalveolates (A), red alga, green alga, plants, fungi, metazoa, and bacteria (B). The conservation was scaled by a heat bar (red) with E-values for putative orthologs of the listed proteins (see Supplementary file 3a). ToxoDB protein numbers (left) and protein (temporary) names (right) are shown. Their sequences were searched for orthologs using the Hidden Markov Model (HMM) (E-value <E−7) and the best ortholog was used for the heatmap analysis in ComplexHeatmap package in R. Parasite fitness scores were shown on the right with the green heat bar for T. gondii (ToxoDB) (Sidik et al., 2016) and the blue bar for P. falciparum (PlasmoDB) (Zhang et al., 2018). The tree dendrogram across the genes were clustered according to the euclidean distance using UPGMA method. The species tree (top) shows phylogenetic relationships and major clades: Theil., Theileria; Babes, Babesia; Crypt., Cryptosporidium; Proteo, Proteobacteria.
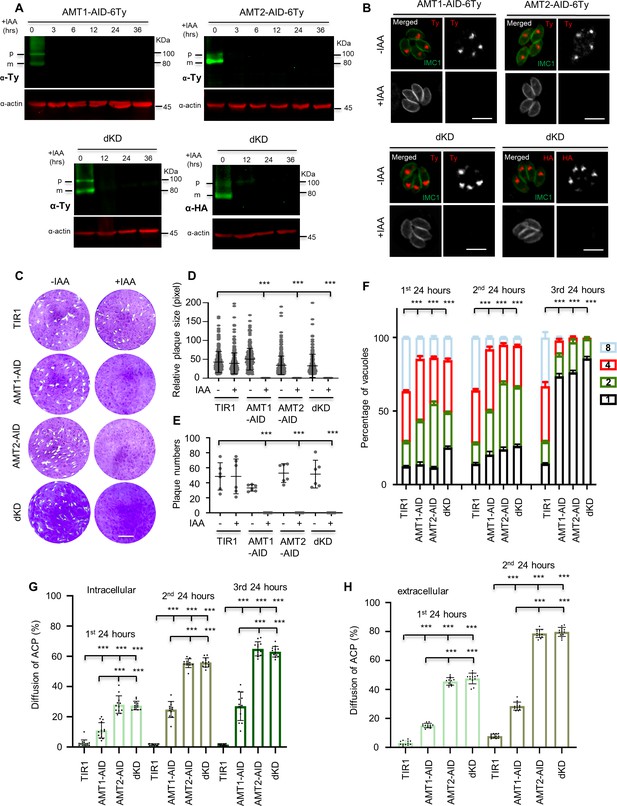
AMT1 and AMT2 are localized at the apicoplast membrane and essential for parasite growth in vitro.
(A) Western blot detection of protein depletion in the IAA-inducible degron (AID) lines. The lines were induced by auxin (IAA) for different hours (hrs) as indicated. Western blots detected immatured (p) and matured (m) forms of the protein fusions in the non-induced lanes. Actin served as the control. (B) Indirect fluorescence assay (IFA) detection of protein depletion in the AID lines. Parasites were grown in IAA for 24 hr, followed by IFA with antibodies against Ty or HA (red) and IMC1 (green). Scale = 5 μm. (C–E) Plaque formation by the TIR1 and AID lines on HFF monolayers in ±IAA for 7 days. Numbers (D) and sizes (E) of the plaques were measured by ImageJ. Scale = 0.5 cm. Two independent experiments with triplicates were performed. Data are shown as a mean ± standard error of the mean (SEM). (F) Parasite replication of the TIR1 and AID lines grown in IAA. The parasites were grown in IAA for 24 hr, followed by scraping, harvesting and infection for the 2nd and 3rd rounds of parasite growth in IAA. The parasites stained with GFP45 were counted in vacuoles (at least 150 vacuoles in each replicate). In comparing to TIR1 (1st, 2nd, and 3rd), p < 0.0001 for the AID lines with 2 and 8 parasites/vacuole in the 1st round, and for the AID lines with 1–8 parasites/vacuole in the 2nd round, p < 0.0001 for the AID lines with 1 and 4 parasites/vacuoles in the 3rd round. (G, H) Acyl carrier protein (ACP) diffusion in the TIR1 and AID lines grown in IAA. Parasites were grown in IAA for the 1st, 2nd, and 3rd lytic cycles, as described in the parasite replication assay (intracellular parasites). Those parasites in the 1st and 2nd round of growth were forced to egress for the same analysis (extracellular parasites). Vacuoles or single parasites were scored (n > 150 for each replicate). Fields/images were selected blind and all parasites/vacuoles were scored on the same fields/images (F–H). Three independent experiments with triplicates were performed (A, B, C, F, G, and H), and representative images were shown for A, B, and C, data are shown as a mean ± SEM with two-way analysis of variance (ANOVA) with Tukey’s multiple comparisons (compared with the TIR1). ***p < 0.0001.
-
Figure 3—source data 1
PDF file containing the uncropped western blot gels for verification of the IAA-inducible degron (AID) fusion lines in Figure 3A.
- https://cdn.elifesciences.org/articles/88866/elife-88866-fig3-data1-v1.zip
-
Figure 3—source data 2
Excel file containing RAW images of plaque assays and scoring results of the parasite growth and apicoplast stability in Figure 3.
- https://cdn.elifesciences.org/articles/88866/elife-88866-fig3-data2-v1.zip
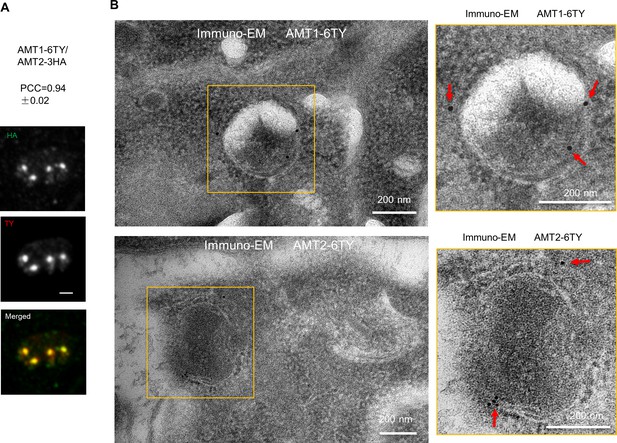
AMT1 and AMT2 are co-localized in the apicoplast.
(A) Confocal images showed co-localization of AMT1-6Ty and AMT2-6HA. PCC was analyzed over the merged fluoresent foci by the NIS Elemental AR system. Data are shown with a mean ± standard deviation (SD; N = 6). (B) Immuno-electron microscopy (immuno-EM) of AMT1-6Ty and AMT2-6Ty. Parasites were fixed, embeded, sliced and incubated sequentially with Ty antibodies and gold particle (15 nm for AMT1 and 10 nm for AMT2) conjugated with anti-mouse antibodies. Scale bars were indicated on the images, and red arrows showed the gold particles at the apicoplast membranes.
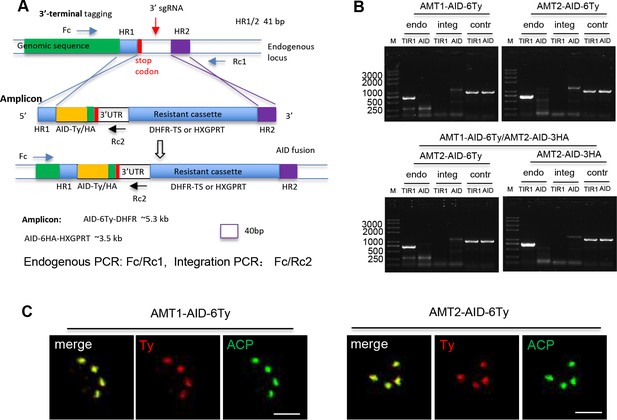
Diagnostic PCR of the AMT1-AID, AMT2-AID, and dKD lines.
(A) Schematic diagram of the generation of the lines using a CRISPR-Cas9 approach. The sgRNA sequences were selected as described previously (Long et al., 2018), and integrated into a pCas9 plasmid using a DNA assembly method. The pCas9-sgRNA 3′ expresses Cas9 and sgRNA, which specifically cleave at the sgRNA region, creating a double strain DNA break (DSB) at the 3′ untranslated region of a specific gene. An amplicon containing an IAA-inducible degron (AID) fragment, a resistant cassette, and homologous regions (HR1 and HR2) at both ends was amplified from a generic plasmid. The HR1 and HR2 targeted the amplicon to integrate at the DSB, resulting in an in-frame integration of the AID fragment at the end of the encoding sequences. The primers were designed for diagnostic PCR for testing endogenous DNA and integration of the amplicon, and a scale bar was added at the bottom of the figure. (B) Diagnostic PCR of the AID lines. Endogenous (endo) and integration (integ) PCRs were performed with pairs of primers Fc/Rc (endo) and Fc/Rc2 (integ) as shown in (A), and the control (contr) PCR was performed with primers for the tubulin promoter region. The product sizes for the endo PCR in both the AID lines were 700 bp, while the sizes of the integ PCR were 1357 bp for AMT1-AID-6Ty, 1407 bp for AMT2-AID-6Ty, and 1326 bp for AMT2-AID-3HA. M, DNA marker. (C) Co-localization of AID-Ty fusions with the apicoplast marker ACP. Parasites were analyzed by IFA using antibodies against Ty (red) and ACP (green), followed by secondary antibodies conjugated with Alexa Fluors. Scale = 5 μm.
-
Figure 3—figure supplement 2—source data 1
PDF file containing DNA gels for diagnostic PCR of the IAA-inducible degron (AID) fusion lines.
- https://cdn.elifesciences.org/articles/88866/elife-88866-fig3-figsupp2-data1-v1.zip
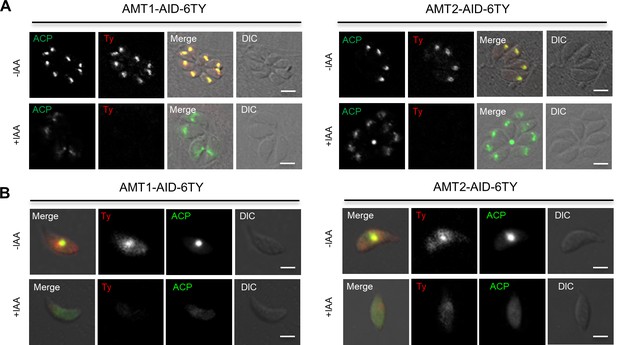
Depletion of AMT1 and AMT2 leads to apicoplast defects and loss of mitochondrial membrane potential in parasites.
Parasites were grown in ±IAA, followed by fixation of intracellular parasites after 24 hr growth (A), or by fixation of extracellular parasites released mechanically after 24 hr growth (B). Indirect fluorescence assay (IFA) was performed with antibodies against epitope tag Ty (red) and apicoplast marker ACP (green). Examples of normal ACP localization, partial diffusion of ACP in the intracellular parasites (A), and a complete diffusion of ACP in the extracellular parasites (B) were shown for the lines of AMT1-AID and AMT2-AID. Scale = 5 μm. DIC, differential interference contrast.
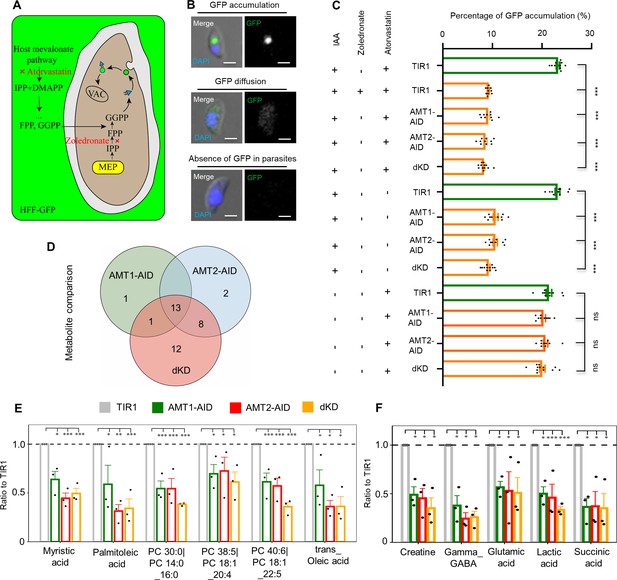
Depletion of AMT1 and AMT2 caused defects of methylerythritol phosphate (MEP) and fatty acid synthesis type II (FASII) pathways.
(A–C) The MEP activity was strongly affected in the IAA-inducible degron (AID) lines. GFP transport to the VAC was assayed by growning parasties in GFP expressing HFF cells (HFF-GFP). LHVS (10 μM) was added to inhibit GFP digestion in the VAC (Dou et al., 2014). The isoprenoid synthesis in host cells and parasites was inhibited by IC50 concentrations of atorvastatin (10 μM) and zoledronate (0.3 μM) (Li et al., 2013), respectively (A). The inhibition over the TIR1 line using two of the drugs was used as a positive control, and representative images were shown for parasites with or withour GFP signal (B). The AID parasites were grown in HFF-GFP cells in the presence of LHVS for 36 hours in total, with the addition of atorvastatin, zoledronate or IAA as stated for different experiments in the figure in the last 12 hours. Parasites were immediately harvested, adhered and scored, as shown in the example images in B. Percentages of parasites with GFP foci were plotted (C). Fields/images were selected blind and all parasites/vacuoles were scored on the same fields/images (C). Three independent experiments with triplicates were performed. Data are shown as a mean ± SEM with one-way ANOVA with Tukey’s multiple comparison. ***, p < 0.0001. Scale = 5 μm. (D-F) Fatty acid synthesis and TCA metabolites were significantly reduced in IAA induced AID parasites. Parasites were grown in IAA for 18 hours and harvested for untargeted metabolomics with GCMS and LCMS (N=3), and metabolites detected were analyzed by Dunnette’s multiple comparison by comparing the AID parasites to the TIR1. Three AID lines shared 13 differential metabolites (D). These shared metabolites included fatty acids and lipids, such as myristic acid, palmitoleic acid, oleic acid, and phosphatidylcholine (PC) (E), and metabolites related to the TCA cycle (F). *, p < 0.05; **, p < 0.001; ***, p < 0.0001.
-
Figure 4—source data 1
Original images and scoring results of the GFP transport assay.
- https://cdn.elifesciences.org/articles/88866/elife-88866-fig4-data1-v1.zip
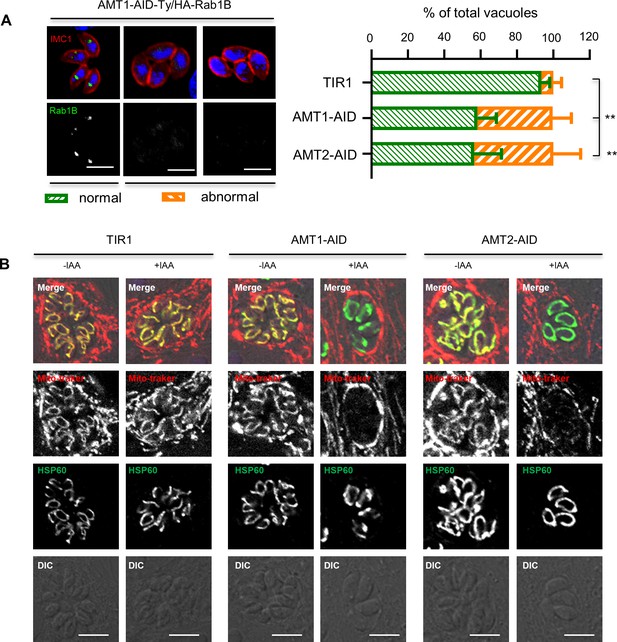
Indirect effect of phenotypes resulted from depletion of AMT1 and AMT2.
(A) Depletion of AMT1 and AMT2 resulted in reduced protein abundance of Rab1B in parasites. Rab1B is a key molecule that regulates endocytic trafficking of GFP vesicle, and whose abundance is regulated by protein prenylation, as demonstrated in our recent study (Wang et al., 2023). Here, depletion of AMT1 and AMT2 resulted in similar phenotype of Rab1B, which mimics that of Rab1B in parasites with inhibited supply of isoprenoids or with knockdown of prenyl-transferase GGT-2. Parasites were grown in IAA for 12 hr and fixed for scoring of vacuoles that contain signal of normal Rab1B (wild-type) or abnormal Rab1B (reduced protein abundance), as shown in the example images. The scoring results were plotted as percentage of parasites with normal and abnormal Rab1B in the populations from three independent experiments with triplicates. Data were shown with mean ± standard deviation (SD) and analyzed by two-way analysis of variance (ANOVA) with Tukey’s multiple comparison. **p < 0.005. Scale = 5 μm. (B) Parasites grown in ±IAA for 18 hr were treated with mitotracker red for 30 min, followed by fixation of the parasites using 4% paraformaldehyde. Indirect fluorescence assay (IFA) was performed with primary antibodies against mitochondrial marker HSP60 and anti-rabbit secondary antibodies conjugated with Alexa Fluor-488 (green). Scale = 5 μm. Three independent experiments were performed with similar outcomes and representative images were shown.
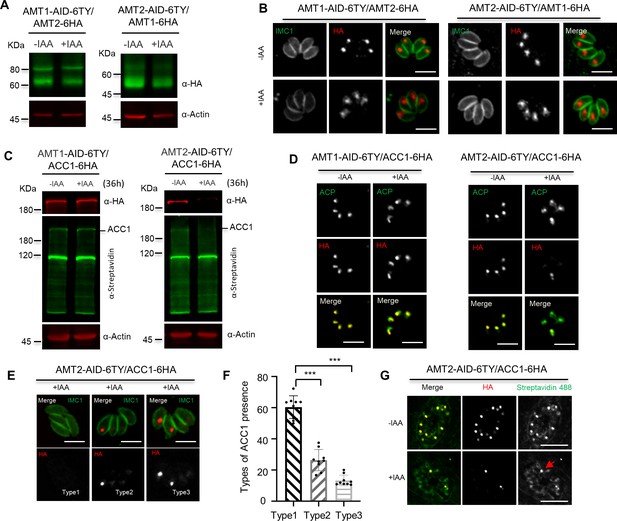
Depletiozn of AMT2 resulted in acetyl-CoA carboxylase 1 (ACC1) instability.
(A, B) No reciprocal effect on protein levels by depletion of AMT1 or AMT2. AMT1 and AMT2 were reciprocally tagged at C-terminus with 6HA in the IAA-inducible degron (AID) lines, and the HA fusions were detected by western blots in parasites in ±IAA for 36 hr (A), and by indirect fluorescence assay (IFA) in parasites in ±IAA for 24 hr (B). IFA was performed with IMC1 (green) and Ty/HA (red). (C, D) ACC1 was lost upon depletion of AMT2-AID. ACC1 was endogenously tagged with 6HA at its C-terminus in the AMT1-AID and AMT2-AID lines, and parasites were grown in IAA for 36 hr for western blots (C) and IFA (D). IFA analyses were performed with anti-ACP and anti-HA antibodies, while western blots were done with anti-HA and streptavidin Li-COR 800CW. Actin served as the control. (E, F) Representative images of ACC1 on IFA in parasites of AMT2-AID induced in IAA for 24 hr. ACC1 (red) was completely lost in parasites of a vacuole (type 1), remained only in one parasite of a single vacuole (type 2), or in two parasites of a single vacuole (type 3) (E). The ACC1 types were scored in AMT2-AID induced in IAA for 24 hr, and expressed as percentages of different types (F). At least 150 parasites were scored in each replicate. Three experiments with triplicates were performed, and data are shown as a mean ± standard error of the mean (SEM) and analyzed by two-way analysis of variance (ANOVA) with multiple comparison, ***p < 0.0001. (G) The remaining ACC1 was still biotinylated. Parasites were grown in ±IAA for 24 hr for IFA analysis with antibodies against HA and streptavidin Alexa Fluor 488. Parasites containing both ACC1-6HA (red) and biotinylated ACC1 (green) was observed. Three independent experiments were performed with similar outcomes, and representative images were shown. Scale = 5 μm.
-
Figure 5—source data 1
PDF file containing the uncropped western blot gels of for strains in Figure 5.
- https://cdn.elifesciences.org/articles/88866/elife-88866-fig5-data1-v1.zip
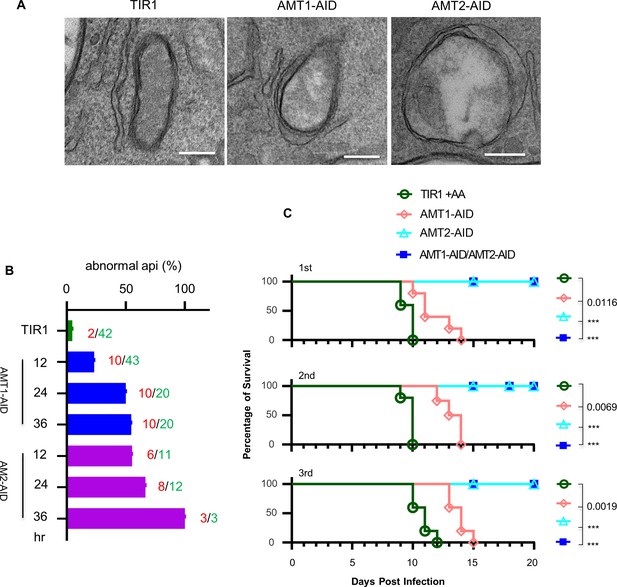
Examination of the apicoplast ultra structures, and mouse virulence of the parasites upon depletion of the IAA-inducible degron (AID) fusions.
(A) Transmission electron microscopy (TEM) observed less electron dense or electron lucent lumen and loosened membranes in the apicoplast in the AID parasites induced in IAA for 24 hr. Representative images were shown. Scale = 200 nm. (B) Quantification of the apicoplast with the lumen less dense or lucent in parasites induced for 12, 24, and 36 hr and TIR in IAA for 36 hr. The TEM examined 300 parasites for three lines (TIR1, AMT1-AID, and AMT2-AID) at each time point, and numbers of abnormal apicoplast (red numbers) and normal apicoplast identified (green numbers) were shown by the columns. The columns express the percentages of abnormal apicoplast in all the identified apicoplasts. (C) Survival curve of BALB/C mice infected with 100 parasites intraperitoneally and treated with IAA for 20 days. The Gehan–Breslow–Wilcoxon test was used to compare differences between the survival curves (the AID line versus TIR1), ***p < 0.0001. The experiment of mouse infection was performed with five mice in each group, and results from three independent experiments (1st, 2nd, and 3rd) were shown together with symbols for the mouse groups.
-
Figure 6—source data 1
Excel file containing the record of mouse infection assay using the IAA-inducible degron (AID) fusion lines.
- https://cdn.elifesciences.org/articles/88866/elife-88866-fig6-data1-v1.zip
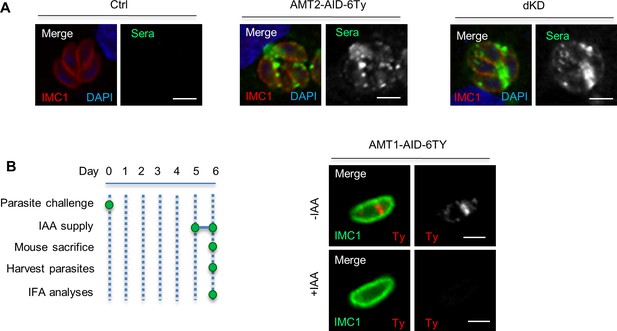
Confirmation of the mice assay by testing mice sera and protein degradation.
(A) Surviving mice were successfully infected with parasites. AMT2-AID and dKD protected mice from lethal toxoplasmosis. All surviving mice were sacrificed at day 20 for extraction of serum, followed by indirect fluorescence assay (IFA) testing of the sera at dilution ratio 1:500 using a standard protocol. Sera from blank mice (Ctrl) without T. gondii infection served as the IFA control. IFA results showed that the sera from surviving mice successfully recognized proteins of the parasitophorous vacuoles of analyzed T. gondii parasites. One representative image was shown for the mouse groups. Scale = 5 μm. (B) The AMT1-AID fusion was efficiently degraded by IAA in mice in 1 day. Mice were intraperitoneally infected by parasites (100 parasites per mouse), followed by growth of parasites in mice for 5 days. The mice were randomly assigned into a control group and an induced group (n = 5 mice). The induced group was orally administered with the drinking water containing IAA for 1 day and intraperitoneally injected with a solution containing IAA on day 5, whereas the control group was supplied with a drinking water and an injection solution without IAA. The mice were then sacrificed on day 6 for extraction of parasites from peritoneal cavities, as demonstrated in the schematic diagram. The parasites were collected and adhered onto poly-lysine-coated coverslips for IFA, which was performed with antibodies against IMC1 (green) and Ty/HA (red). IFA showed that the IAA-inducible degron (AID) fusion was efficiently degraded by IAA in mice within 1 day, and one representative image for each line infection was shown. Scale = 5 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Toxoplasma gondii) | As listed in Supplementary file 2 | ToxoDB release 53 | Supplementary file 2 | |
| Strain, strain background (T. gondii) | T. gondii RHΔhxgprtΔku80; RHΔhxgprtΔku80/TIR1 and Supplementary file 1b | Generous gifts from Prof. Vern Carruthers and Prof. David Sibley PMID:19218426 PMID:28465425 | See Materials and methods | |
| Strain, 6- to 8-week-old femal, Balb/C (Mus musculus) | Balb/C | Division of Experimental Animals, Beijing University | Strain code 028 | |
| Recombinant protein | Supplementary file 2c | This paper | Supplementary file 2c | See Materials and methods |
| Softwares, algorithm | PRISM, version 8.0 | http://www.graphpad.com | GraphPAD v8.0 RRID:SCR_002798 | |
| Softwares, algorithm | InterPro:protein sequence analysis & classification | https://www.ebi.ac.uk/interpro/ | InterPro RRID:SCR_006695 | |
| Softwares, algorithm | Toxoplasma Genomics Resource | http://toxodb.org/toxo/ (Alvarez-Jarreta et al., 2024) | ToxoDB release 53 | |
| Softwares, algorithm | Eukaryotic Pathogen Genomics Resources | https://veupathdb.org/veupathdb (Alvarez-Jarreta et al., 2024) | VEuPathDB | |
| Softwares, algorithm | MetaboAnalyst 5.0 | https://www.metaboanalyst.ca (Pang et al., 2021) PMID:34019663 | MetaboAnalyst | |
| Softwares, algorithm | jackhmmer | http://hmmer.org/ (Prakash et al., 2017) PMID:29220076 | HMMER v3.3.2 | |
| Softwares, algorithm | trimAL v1.2.rev57 | https://vicfero.github.io/trimal/ (Capella-Gutiérrez et al., 2009) PMID:19505945 | trimAL v1.2.rev57 | |
| Softwares, algorithm | TBtools | https://github.com/CJ-Chen/TBtools (Chen and Zhao, 2023) PMID:37740491 | ||
| Softwares, algorithm | R v4.2.0 | https://www.r-project.org/ | RRID:SCR_001905 | |
| Softwares, algorithm | ComplexHeatmap | https://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html (Gu et al., 2016) PMID:27207943 | ||
| Softwares, algorithm | ChiPlot | https://www.chiplot.online/ | ||
| Softwares, algorithm | Mafft v7.490 | https://mafft.cbrc.jp/alignment/software/ (Katoh et al., 2019) PMID:28968734 | ||
| Chemical compound, drug | Pyrimethamine | Sigma-Aldrich | Cat#46706 | 3 µM |
| Chemical compound, drug | Mycophenolic acid | Sigma-Aldrich | Cat#M5255 | 25 µg/ml |
| Chemical compound, drug | Xanthine | Sigma-Aldrich | Cat#X4002 | 25 µg/ml |
| Chemical compound, drug | 3-Indoleacetic acid (auxin) | Sigma-Aldrich | Cat#I2886 | 500 µM |
| Chemical compound, drug | D-biotin | Sigma-Aldrich | Cat#B4639 | 500 µM |
| Chemical compound, drug | 4% paraformaldehyde solution | Solarbio PMID:37108334 PMID:36813769 | Car#P1110 | |
| Antibody | Mouse anti-Ty (BB2) | In house hybridoma; gift from Prof. Philippe Bastin | 1:100 | |
| Antibody | Rabbit polyclonal anti-HA (SG77) | Thermo Fisher Scientific | Cat#71-5500 | 1:500 |
| Antibody | Mouse monoclonal anti-HA | BioLegend | Cat#901501 | 1:500 |
| Antibody | Rabbit polyclonal anti-GAP45 | Home-made PMID:36813769 | Polyclonal | 1:500 |
| Antibody | Rabbit polyclonal anti-actin | Home-made PMID:36813769 | Polyclonal | 1:1000 |
| Antibody | Rabbit polyclonal anti-IMC1 | Home-made PMID:36813769 | Polyclonal | 1:1000 |
| Antibody | Rabbit polyclonal anti-ACP | Home-made PMID:36813769 | Polyclonal | 1:1000 |
| Antibody | Mouse monoclonal anti-GFP | Thermo Fisher Scientific | Cat#MA5-15349 | 1:500 |
| Antibody | Streptavidin Alexa Fluor-488 conjugate | LICOR | Cat#926-3230 | 1:2000 |
| Antibody | Alexa Fluor 488 Goat anti-mouse IgG (H+L) | Thermo Fisher Scientific | Cat#A-11029 | 1:2000 |
| Antibody | Alexa Fluor 568 Goat anti-mouse IgG (H+L) | Thermo Fisher Scientific | Cat#A-11031 | 1:2000 |
| Antibody | Alexa Fluor 568 Goat anti-rabbit IgG (H+L) | Thermo Fisher Scientific | Cat#A-11036 | 1:2000 |
| Antibody | Alexa Fluor 488 Goat anti-rabbit IgG (H+L) | Thermo Fisher Scientific | Cat#A-11034 | 1:2000 |
| Antibody | IRDye 800CW Goat anti-mouse IgG (H+L) | LI-COR | Cat#926-32210 | 1:2000 |
| Antibody | IRDye 800CW Goat anti-rabbit IgG (H+L) | LI-COR | Cat#926-32211 | 1:2000 |
| Antibody | IRDye 680CW Goat anti-rabbit IgG (H+L) | LI-COR | Cat#926-68071 | 1:2000 |
| Antibody | IRDye 680CW Goat anti-mouse IgG (H+L) | LI-COR | Cat#926-68070 | 1:2000 |
| Antibody | IRDye 800CW Streptavidin | LI-COR | Cat#926-32230 | 1:2000 |
| Commercial assay or kit | pEASY-Basic Seamless Cloning and Assembly Kit | Transgene | CU201-02 | |
| Commercial assay or kit | Pierce Streptavidin Magnetic Beads | Thermo Fisher Scientific | Cat#88816 | |
| Commercial assay or kit | ProLong Gold Antifade Mountant without DAPI | Thermo Fisher Scientific | Cat#36930 | |
| Commercial assay or kit | ProLong Gold Antifade Mountant with DAPI | Thermo Fisher Scientific | Cat#36931 | |
| Cell line (include species here) | Human Foreskin Fibroblasts (human) | ATCC | Cat#SCRC-1041 | |
| Others | Fetal bovine serum (solution) | Transgene PMID:37108334 PMID:36813769 | FS201-02 | |
| Others | Dulbecco’s modified Eagle medium (DMEM) (powder) | Thermo Fisher Scientific PMID:37108334 PMID:36813769 | Cat#12800-082 | |
| Others | Penicillin–streptomycin (100×) | Solarbio PMID:37108334 PMID:36813769 | P1400 | |
| Others | Glutamine (100×) | Solarbio PMID:37108334 PMID:36813769 | G0200 |
Additional files
-
Supplementary file 1
Candidate selection and reagents of parasite lines, plasmids, and primers used in the study.
(a) Selection of candidates from previous studies of HyperLOPIT AND PfACP-BirA. (b) Lines used in this study. (c) Plasmids used in this study. (d) Primers used in this study.
- https://cdn.elifesciences.org/articles/88866/elife-88866-supp1-v1.xlsx
-
Supplementary file 2
Analysis of candidates identified from our TurboID proteomics or other sources.
(a) Minimally processed mass-spectrometry datasets of the APT1-TurboID fusion line and the parental line. (b) Volcano analysis of mass-spectrometry datasets from APT1-TurboID and the parental line. (c) List of known apicoplast proteins on the volcano plotting. (d) List of transmembrane domain (TMD) proteins identified by APT1-TurboID and ordered from high numbers to low numbers. (e) Merged candidate list from three datasets of hyperLOPIT, ACP-BirA, and APT1-TurboID. (f) Bioinformatic analysis of discovered transmembrane proteins in this study.
- https://cdn.elifesciences.org/articles/88866/elife-88866-supp2-v1.xlsx
-
Supplementary file 3
Identification of orthologs of apicoplast proteins in diverse taxa.
(a) Identification of orthologs of novel apicoplast proteins and known proteins in chromalveolates, red algae, green algae, plants, fungi, metazoa, and bacteria. (b) Orthologs of AMT1 and AMT2 in selected species of apicomplexans.
- https://cdn.elifesciences.org/articles/88866/elife-88866-supp3-v1.xlsx
-
Supplementary file 4
Analysis of proteins lines by gas chromatography–mass spectrometry (GC–MS) and liquid chromatography–mass spectrometry (LC–MS).
(a) Minimally processed datasets of metabolites detected by GC–MS and LC–MS in TIR1, AMT1-AID, AMT2-AID, and dKD lines induced by auxin for 18 hr. (b) Analysis of metabolites detected by GC–MS and LC–MS in the lines of TIR1, AMT1-AID, AMT2-AID, and dKD induced by auxin for 18 hr.
- https://cdn.elifesciences.org/articles/88866/elife-88866-supp4-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/88866/elife-88866-mdarchecklist1-v1.docx





