An unconventional gatekeeper mutation sensitizes inositol hexakisphosphate kinases to an allosteric inhibitor
Figures
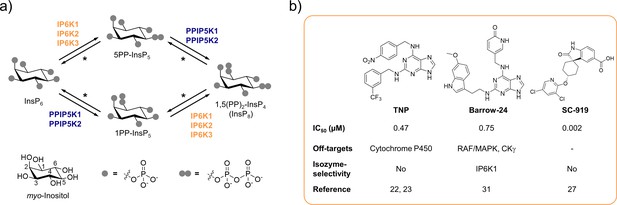
The inositol pyrophosphate pathway is a therapeutically relevant target for small molecule inhibitors.
(a) Simplified inositol pyrophosphate metabolism with a focus on kinases. PPIP5K: inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase. *PP-InsP dephosphorylation is catalyzed by nudix hydrolases DIPP1/2α/2β/3 (Lonetti et al., 2011; Hua et al., 2003; Saiardi et al., 2000). (b) Chemical structures and properties of selected IP6K inhibitors: widely used TNP, isozyme-selective Barrow-24, and potent inhibitor SC-919.
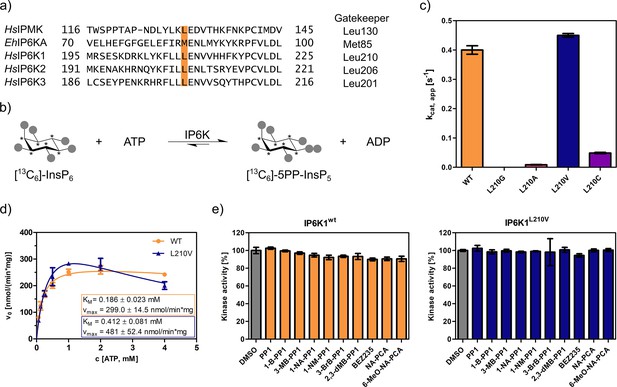
An unusual valine gatekeeper mutant retains catalytic activity.
(a) Sequence alignment of InsP kinases to identify the gatekeeper position of human inositol hexakisphosphate kinases (IP6Ks). The gatekeeper residue is highlighted in orange. (b) Kinase reaction of IP6Ks using uniformly 13C-labeled InsP6 as substrate. Asterisks indicate 13C-labeled positions. The conversion to 5PP-InsP5 was followed by an established spin-echo difference NMR method (Harmel et al., 2019; Riemer et al., 2021). (c) Catalytic activity of IP6K gatekeeper mutants indicated by apparent turnover numbers kcat, app. (d) Michaelis–Menten graphs for IP6K1wt and IP6K1L210V. (e) Screening of established analog-sensitive kinase inhibitors (Figure 2—figure supplement 4) at 10 µM concentration against IP6K1wt and IP6K1L210V using the NMR assay. All data points were measured in independent triplicates and error bars represent the standard deviation.
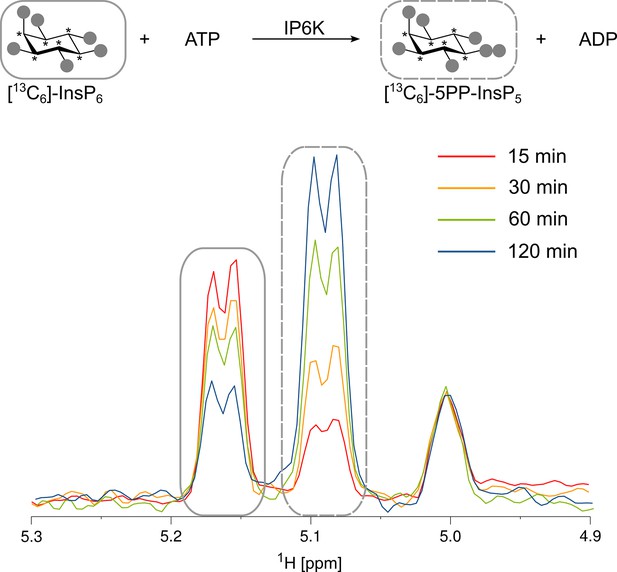
Kinase reaction of inositol hexakisphosphate kinases (IP6Ks) using fully 13C-labeled InsP6 as a substrate.
The conversion to 5PP-InsP5 was followed by the established spin-echo difference NMR method (Harmel et al., 2019). The peaks corresponding to substrate and product are framed with solid or dashed rectangles, respectively. Asterisks indicate 13C-labeled positions.
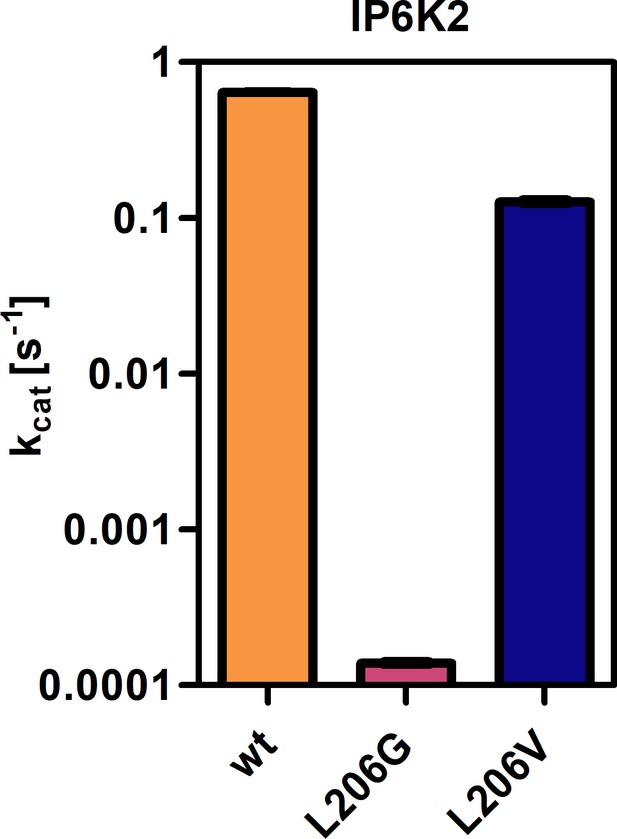
Catalytic activities of IP6K2 wild-type (WT) and gatekeeper mutants as indicated by the apparent turnover number.
All samples were measured in independent triplicates and error bars represent standard deviation.
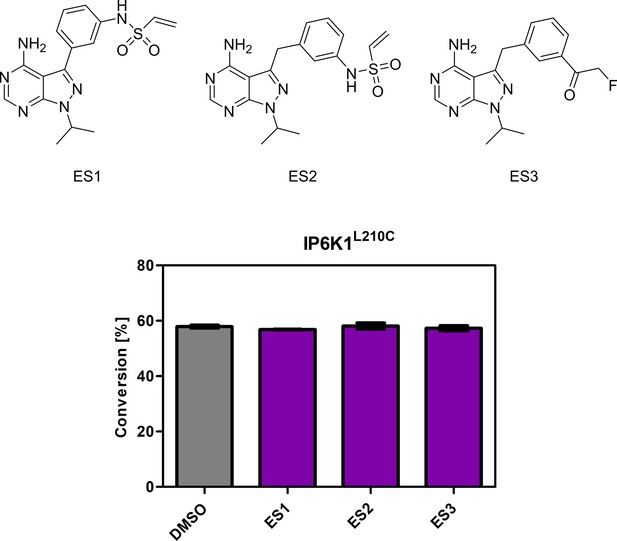
Screening of established electrophile-sensitive kinase inhibitors at 10 µM concentration against IP6K1L210C using the NMR assay.
All compounds were measured in independent triplicates and error bars represent standard deviation.
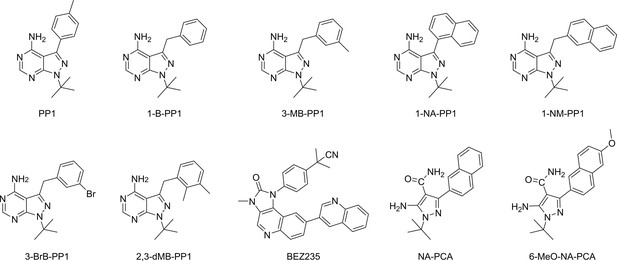
Chemical structures of established analog-sensitive kinase inhibitors screened against IP6K1L210V.
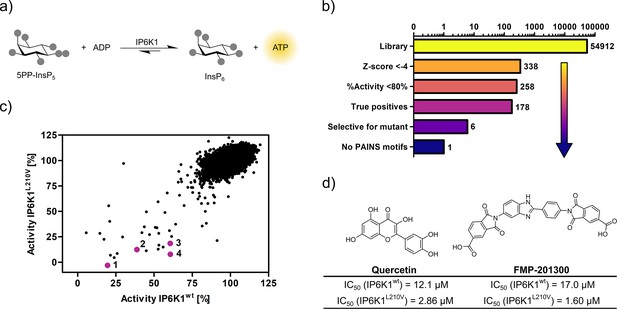
High-throughput screening provides gatekeeper mutant-selective hit compound.
(a) ATP synthase reaction used for the high-throughput screen. The generation of ATP was monitored using a luminescence-based Kinase-Glo assay. (b) Reduction of putative hits by applying specific thresholds and manual selection. (c) Scatter plot comparing the potency of compounds measured in the primary screen against IP6K1wt and IP6K1L210V. Hits highlighted in magenta represent known IP6K inhibitors, or promising hits. 1: 6-hydroxy-dl-DOPA, 2: FMP-201300, 3: myricetin, 4: quercetin. (d) Examples of true positive hits that display selectivity for IP6K1L210V, including known IP6K inhibitor quercetin and newly discovered FMP-201300.
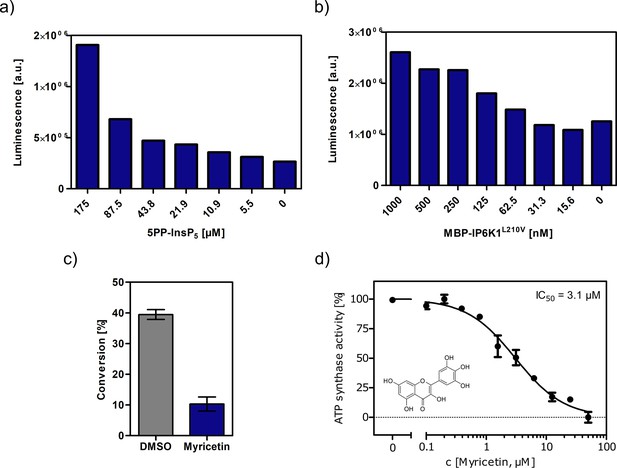
Optimization of assay conditions for high-throughput screening.
(a, b) Serial dilution of substrate and protein in single measurements to determine suitable concentrations for the high-throughput screen using the Kinase-Glo assay. (c) Substrate conversion at optimized reaction conditions measured by NMR. 10 µM myricetin was used as positive control. (d) IC50 curve of myricetin against IP6K1L210V in reverse reaction measured by NMR. All concentrations were measured in independent triplicates and error bars represent standard deviation.
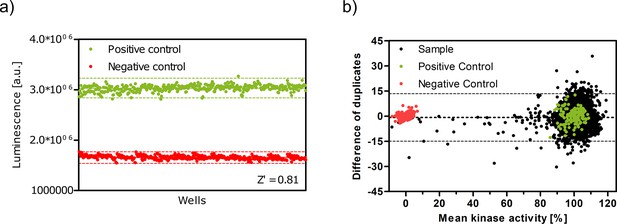
Z' plate measurement and Bland-Altman plot indicate high assay quality.
(a) Z′-plate with positive and negative controls to assess the assay quality and high-throughput screen viability. Dashed lines indicate the ± 3*standard deviation values. (b) Bland–Altman plot of pilot screen. The dotted line indicates the mean of the differences and dashed lines represent the limits of agreement (±14.2%).

FMP-201300 appears to have very specific interactions with the kinase.
(a) IC50 curves of FMP-201300 against IP6K1wt and IP6K1L210V. (b) IC50 curves of FMP-201300 against IP6K2wt and IP6K2L210V. 100% activity corresponds to the DMSO control and 0% indicates no substrate conversion. (c) Structures of FMP-201300 analogs and their inhibitory activities against IP6K1L210V at 10 µM concentration. (d) Lineweaver–Burk plot of FMP-201300 against IP6K1L210V at two different inhibitor concentrations. The plotted lines for the DMSO control and two different inhibitor concentrations intersect almost precisely on the x-axis, indicating no change in KM value and a decrease in vmax upon inhibition (e) IC50 curves of FMP-201300 against IP6K1L210V at two different InsP6 concentrations. All data points were measured in independent triplicates and error bars represent the standard deviation.
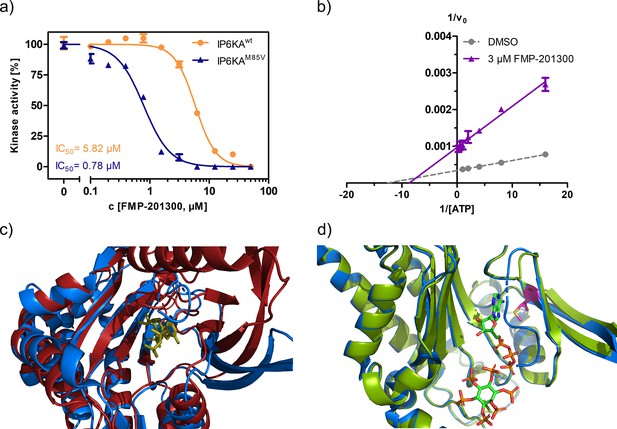
Characterization of the interaction between FMP-201300 and EhIP6KA.
(a) IC50 curves of FMP-201300 against IP6KAwt and IP6KAM85V. (b) Lineweaver–Burk plot of FMP-201300 against IP6KAwt. All points were measured in independent triplicates and error bars represent standard deviation. (c) Zoom-in on the structural alignment of EhIP6KA (blue) bound to ATP (olive) (PDB: 4O4F) and the AlphaFold structure model of IP6K1 (red) (Q92551) (Jumper et al., 2021). (d) Zoom-in on the structural alignment of published EhIP6KAwt crystal structure bound to ATP and InsP6 (blue, PDB: 4O4F), and IP6KAM85V crystal structure (green, this study, PDB: 8OMI, bound ATP omitted for clarity). The gatekeeper residue is highlighted in magenta.
-
Figure 4—figure supplement 1—source data 1
Data collection and structure refinement statistics of EhIP6KAM85V crystallization.
- https://cdn.elifesciences.org/articles/88982/elife-88982-fig4-figsupp1-data1-v1.docx
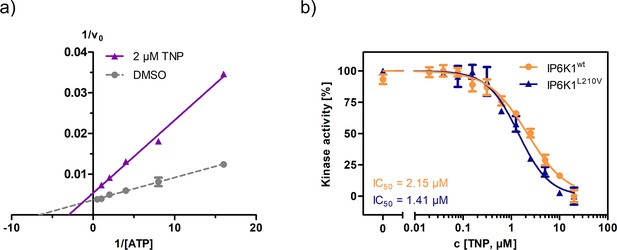
Established pan-IP6K inhibitor TNP displays ATP-competitive mechanism and no selectivity towards the valine gatekeeper mutant.
(a) Lineweaver–Burk plot of TNP against IP6K1wt. (b) IC50 curves of TNP against IP6K1wt and IP6K1L210V. All points were measured in independent triplicates and error bars represent standard deviation.
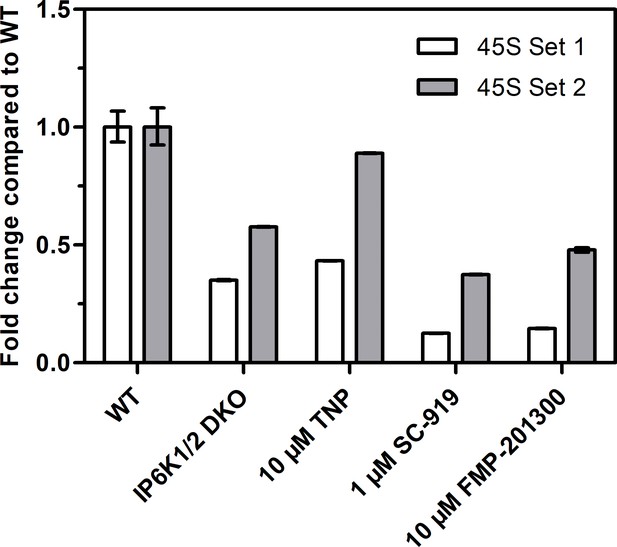
Reverse transcription quantitative PCR analysis to measure 45S pre-rRNA transcript levels using two different primer sets.
Values indicate the fold change in transcript levels in IP6K1/2 double knockout (DKO) cells, or TNP-, SC-919-, or FMP-201300-treated HCT116 cells compared to HCT116 WT cells. The fold change was calculated using the ΔΔct method and independent technical triplicates.
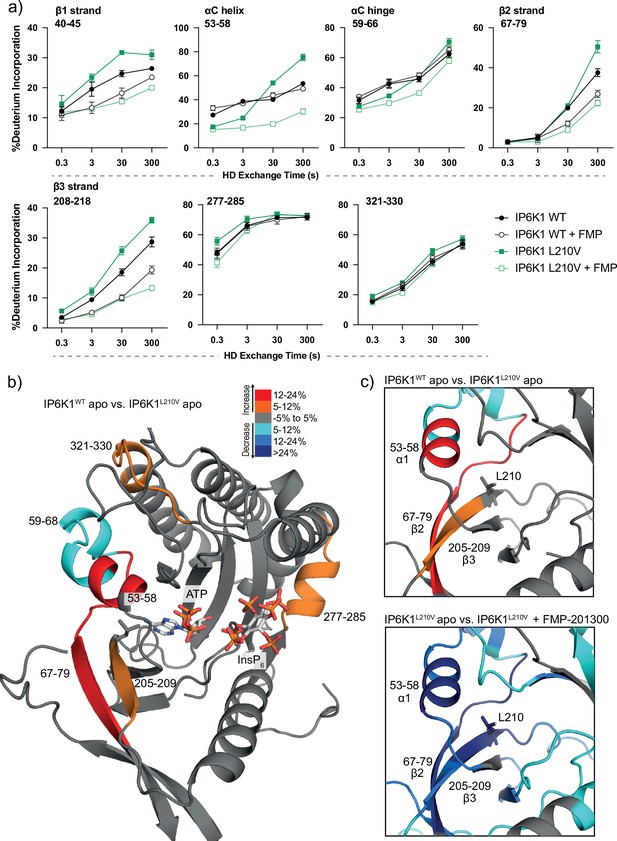
Hydrogen deuterium exchange mass spectrometry (HDX-MS) reveals increased flexibility in the β2–β3 strands and αC helix induced by the IP6K1L210V mutation that sensitize the enzyme to FMP-201300.
(a) HDX differences in IP6K1. Time course of deuterium incorporation for a selection of peptides. Raw data can be found in the Supplementary file 1. (b) Overall HDX-MS changes in deuterium incorporation induced by the IP6K1L210V mutation. Differences in deuterium exchange rates mapped on a model of IP6K1wt (AlphaFold structure prediction Q92551 with ATP and InsP6 from EhIP6KA docked in the active site (PDB: 4O4F)). Peptides that showed significant differences in HDX and met the cut-offs were included (>6% deuterium incorporation and 0.5 Da with an unpaired Student’s t-test of p < 0.05). Strongly disordered regions and regions of low per-residue confidence scores (pLDDT) were omitted for clarity. (c) A magnified view of gatekeeper region included the β2–β3 strands and αC helix. The same regions that show increased flexibility in the IP6K1L210V mutant (top panel) also undergo large decreases in exchange upon binding to FMP-201300 (bottom panel). This increased flexibility likely allows accommodation of the inhibitor and ATP.
-
Figure 5—source data 1
Hydrogen deuterium exchange mass spectrometry raw data.
- https://cdn.elifesciences.org/articles/88982/elife-88982-fig5-data1-v1.xlsx
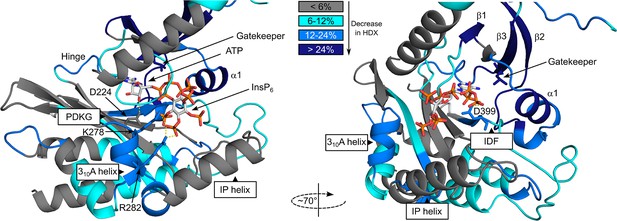
Hydrogen deuterium exchange mass spectrometry (HDX-MS) shows FMP-201300 binding to IP6K1L210V leads to decreases that correspond to an allosteric mechanism of action.
Differences in deuterium exchange rates mapped on a model of IP6K1wt (AlphaFold structure prediction Q92551 with ATP and InsP6 from EhIP6KA docked in the active site (PDB: 4O4F)). Peptides that showed significant differences in HDX and met the cut-offs were included (>6% deuterium incorporation and 0.5 Da with an unpaired Student’s t-test of p < 0.05). Differences in deuterium incorporation >6%,>12%, and>24% compared to the apo enzyme are colored in teal, blue, and navy, respectively. Gray regions had no altered deuterium uptake. Strongly disordered regions and regions of low per-residue confidence scores (pLDDT) were omitted for clarity.
Additional files
-
Supplementary file 1
Hit compounds selective for IP6K1L210V.
FMP-201300, highlighted in green, is the most promising compound as it is not displaying any PAINS motifs, has no known inhibitory activities, and is neither redox-active nor cytotoxic. All values are for IP6K1L210V if not otherwise indicated.
- https://cdn.elifesciences.org/articles/88982/elife-88982-supp1-v1.xlsx
-
Supplementary file 2
Cytotoxicity of selected hits indicated by cell survival after 72 hr incubation at 10 µM concentration.
All compounds were measured in independent triplicates and errors represent standard deviation. Redox activity of selected compounds measured via a photometric surrogate assay. Reactions contained 50 mM HEPES pH 7.5, 50 mM NaCl, 200 mM DTT, and 20 µM resazurin, and were incubated for 1 hr at 37°C.
- https://cdn.elifesciences.org/articles/88982/elife-88982-supp2-v1.xlsx
-
Supplementary file 3
Primers used for ligation-independent cloning and site-directed mutagenesis.
- https://cdn.elifesciences.org/articles/88982/elife-88982-supp3-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/88982/elife-88982-mdarchecklist1-v1.pdf





