Vacuolar H+-ATPase determines daughter cell fates through asymmetric segregation of the nucleosome remodeling and deacetylase complex
Figures
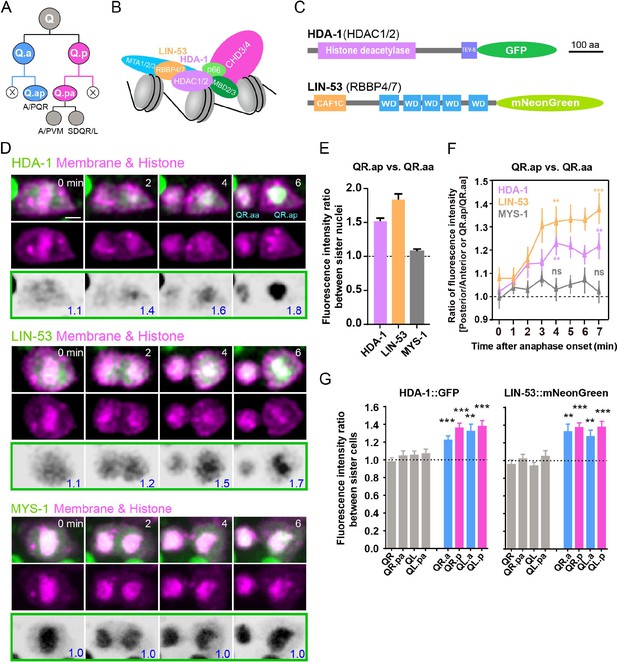
Asymmetric segregation of nucleosome remodeling and deacetylase (NuRD) during asymmetric cell divisions (ACDs) of C. elegans Q neuroblast.
(A) Schematic of the Q neuroblast lineages. QL or QR neuroblast each generates three neurons and two apoptotic cells (Q.aa/Q.pp, X). QL produces PQR, PVM, and SDQL. QR produces AQR, AVM, and SDQR. (B) A model of the NuRD complex composition (Bracken et al., 2019; Lai and Wade, 2011). (C) Protein domain structure of the GFP-tagged HDAC1/2 (HDA-1) or mNeonGreen-tagged RBBP4/7 (LIN-53). CAF1C: histone-binding protein RBBP4 or subunit C of CAF1 complex; WD: WD40 repeat tryptophan-aspartate domain. Scale bar: 100 amino acids. (D) Representative images of endogenous HDA-1::GFP, LIN-53::mNeonGreen and overexpressed MYS-1::GFP during ACDs of QR.a. In each panel, the top row shows merged images, the middle row shows mCherry-tagged plasma membrane and histone, and the bottom row shows inverted fluorescence images of GFP/mNeonGreen. The anterior of the cell is on the left. The GFP/mNeonGreen fluorescence intensity ratios between posterior and anterior chromatids, and between QR.ap and QR.aa nuclei, are shown in blue at the lower-right corner of inverted fluorescence images. Other frames are in Figure 3A, and the full movies are in Videos 3–5. Scale bar: 2 µm. (E) Quantification of HDA-1::GFP, LIN-53::mNeonGreen and MYS-1::GFP fluorescence intensity ratio between QR.ap and QR.aa nuclei. Data are presented as mean ± SEM. N = 10–12. (F) Quantification of HDA-1::GFP (magenta), LIN-53::mNeonGreen (orange), and MYS-1::GFP (gray) fluorescence intensity ratios between the posterior and anterior half of QR.a or between QR.ap and QR.aa. Anaphase onset is defined as the last frame without chromatids segregation. Data are presented as mean ± SEM. N = 10–12. Statistical significance is determined by a one-sample t-test, with 1 as the theoretical mean. **p<0.01, ***p<0.001, ns: not significant. (G) Quantification of HDA-1::GFP (left) and LIN-53::mNeonGreen (right) fluorescence intensity ratios between the large and small daughters of cells shown on the X-axis. Data are presented as mean ± SEM. N = 7–14. Statistical significance is determined by a one-sample t-test, with 1 as the theoretical mean. **p<0.01, ***p<0.001.
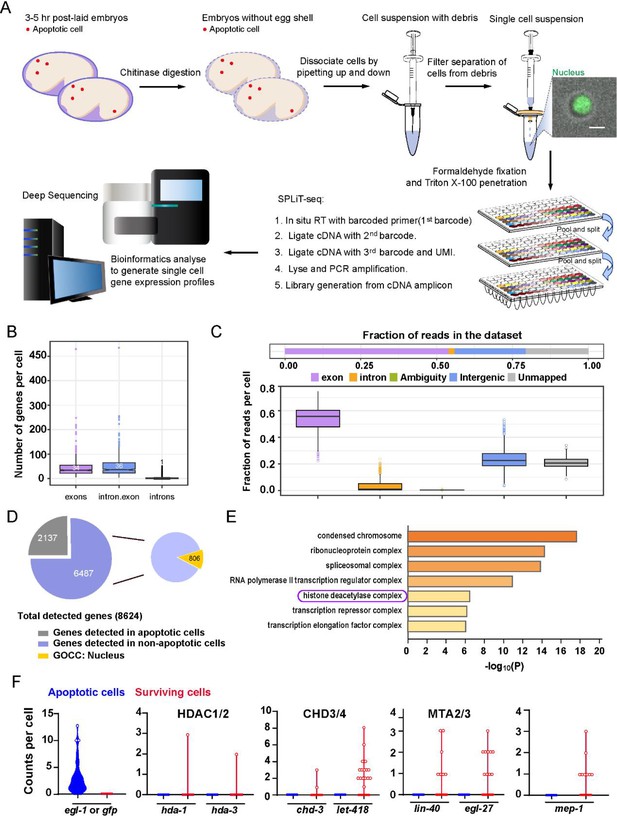
Single-cell sequencing of the C. elegans embryonic cells.
(A) Schematic of the single-cell SPLiT-seq workflow for C. elegans embryonic cells. (B) Number of genes detected per cell is generated from exon, intron, and exon plus intron mapped reads. Each dot represents a cell, and each box represents the median and first and third quartiles per group. (C) The distribution of reads into different mapping feature categories per cell. Each dot represents a cell, and each box represents the median and first and third quartiles. (D) The left pie chart shows the proportion of genes detected in SPLiT-seq data from different cell types. The numbers within the pie chart indicate the number of genes. The right pie chart shows that 806 out of 6487 genes, which were not detected in the egl-1-positive cells, show enrichment for the nuclear GO terms in the Gene Ontology Cellular Components (GOCC) analysis. See also Supplementary file 1. (E) Bar plot ranking of representative GOCC terms with a p-value lower than 10–6 of 806 genes localized to the nucleus. The ‘histone deacetylase complex’ GOCC term includes hda-1, chd-3, lin-40, and egl-27, which encode nucleosome remodeling and deacetylase (NuRD) subunits. (F) Violin plots show counts per cell for egl-1/Pegl-1::gfp and NuRD component-encoding genes in apoptotic cells and surviving cells.

Asymmetric segregation of overexpressed nucleosome remodeling and deacetylase (NuRD) during asymmetric cell division (ACD) of QR.a.
Fluorescence time-lapse images of overexpressed GFP-tagged NuRD subunits and mCherry-tagged plasma membrane and histone during asymmetric divisions of QR.a cells. In each panel, the top row shows merged images, the middle row shows mCherry-tagged plasma membrane and histone, and the bottom row shows inverted fluorescence images of GFP. Scale bar: 2 µm. See also Videos 1 and 2.
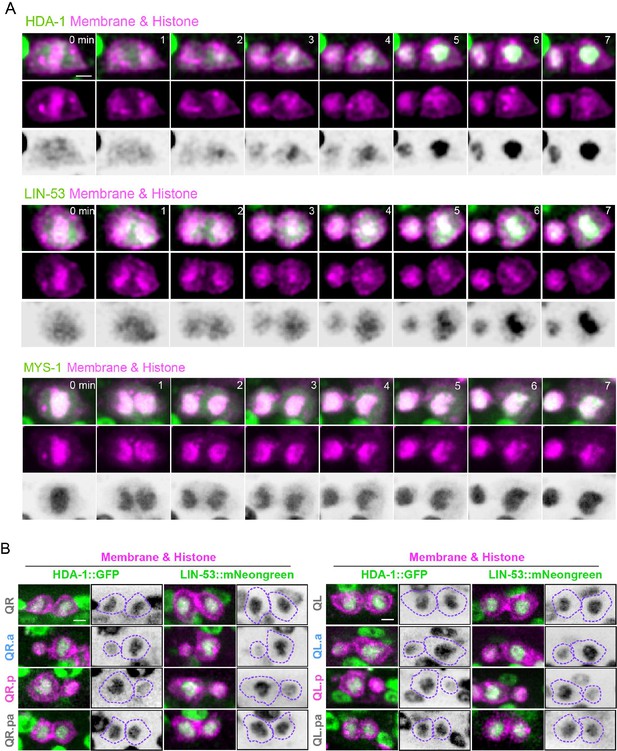
Asymmetric segregation of endogenous nucleosome remodeling and deacetylase (NuRD) during asymmetric cell divisions (ACDs) of Q cells.
(A) Representative images of HDA-1::GFP (top), LIN-53::mNeonGreen (middle), and MYS-1::GFP (down) during QR.a division. The anterior of the cell is on the left. In each panel, the top row shows merged images, the middle row shows mCherry-tagged plasma membrane and histone, and the bottom row shows inverted fluorescence images of GFP/mNeonGreen. Scale bar: 2 µm. See also Videos 3–5. (B) Representative fluorescence images show HDA-1::GFP (left) and LIN-53::mNeonGreen (right) in daughters of indicated Q cells. Inverted fluorescence images of GFP GFP/mNeonGreen signal are on the right of merged images. Dotted purple lines show cell peripheries. The anterior of the cell is toward the left. Scale bar: 2 µm.
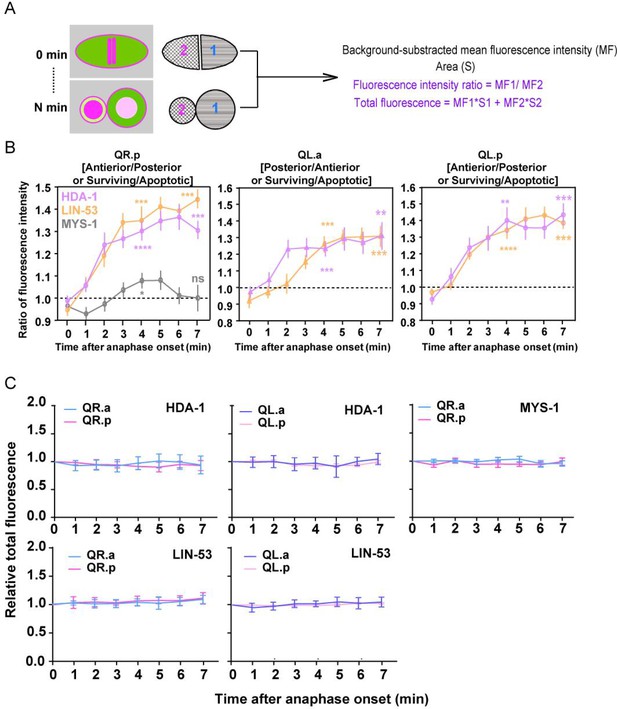
Quantifications of asymmetric nucleosome remodeling and deacetylase (NuRD) segregation during asymmetric cell division (ACD).
(A) Schematics of the fluorescence quantification method. (B) Quantification of the fluorescence intensity ratio (MF1/MF2) changes of HDA-1, LIN-53, and MYS-1 in dividing Q cells. Data are presented as mean ± SEM. N = 10–12. Statistical significance is determined by a one-sample t-test, with 1 as the theoretical mean. *p<0.05, **p<0.01, ***p<0.001, ***p<0.0001, ns: not significant. (C) The relative total fluorescence of HDA-1, LIN-53, and MYS-1 during ACDs in Figure 1F and B. The total fluorescence (MF1*S1+MF2*S2) at each point was normalized to that at time point 0 min. Data are presented as mean ± SEM. N = 10–12.
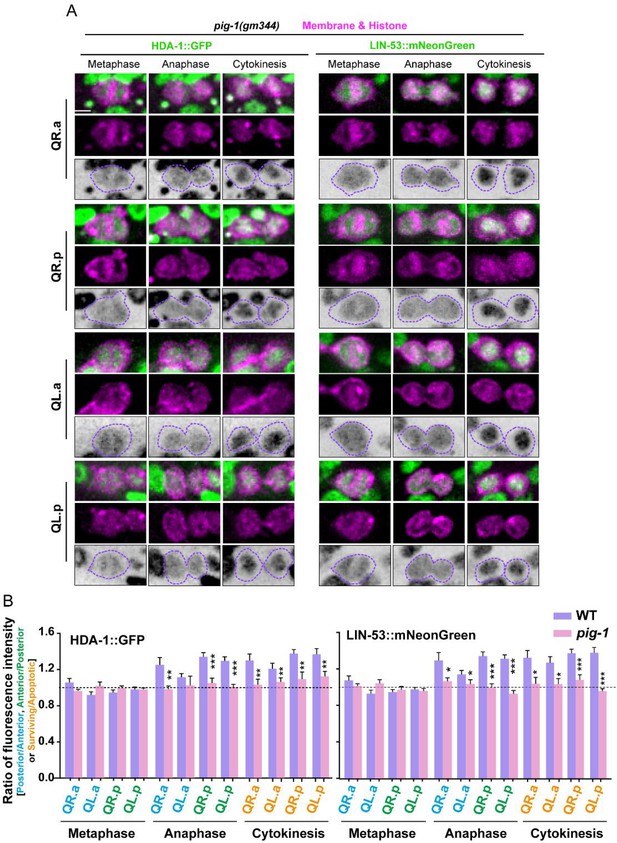
Symmetric nucleosome remodeling and deacetylase (NuRD) segregation in pig-1 mutant.
(A) Representative images of HDA-1::GFP (left) and LIN-53::mNeonGreen (right) during asymmetric cell divisions (ACDs) of Q cells in pig-1 (gm344) mutant. In each panel, the top row shows merged images, the middle row shows mCherry-tagged plasma membrane and histone, and the bottom row shows inverted fluorescence images of GFP/mNeonGreen. Anterior of the cell is left. Scale bar: 2 µm. See also Videos 6 and 7. (B) Fluorescence intensity ratios of HDA-1::GFP (left) and LIN-53::mNeonGreen (right) between the surviving and apoptotic daughters of Q cells in WT or pig-1 (gm344) mutants. Data are presented as mean ± SEM. N = 9–19. Statistical significance is determined by Student’s t-test. *p<0.05, **p<0.01, ***p<0.001.
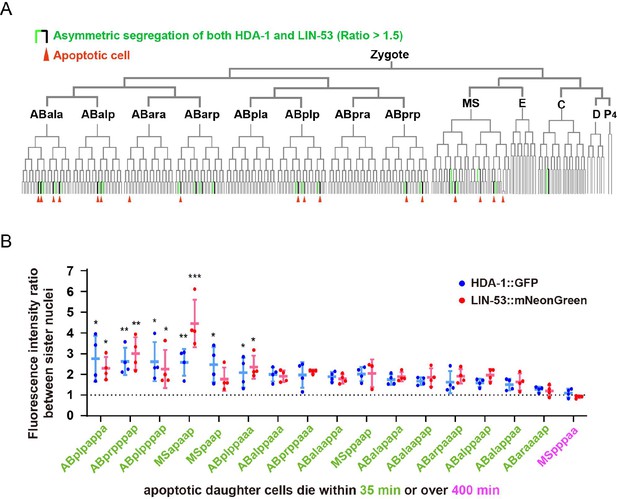
Nucleosome remodeling and deacetylase (NuRD) asymmetry in C. elegans embryonic cell lineages.
(A) The tree visualization depicts the segregation of HDA-1::GFP and LIN-53::mNeonGreen between sister cells during embryonic development. In this tree structure, vertical lines represent cells and horizontal lines denote cell divisions. Green vertical lines highlight cells with higher nuclear HDA-1::GFP and LIN-53::mNeonGreen fluorescence intensity than their apoptotic sister cells (average fluorescence intensity ratio between sister cell nuclei >1.5). Red arrowheads point to apoptotic cells. The placement of cells within the tree follows the Sulston nomenclature. See also Supplementary file 2. (B) Quantifications of HDA-1::GFP and LIN-53::mNeonGreen fluorescence intensity ratios between nuclei of live daughter cells and their apoptotic sister cells. The lineage names of 17 cells that divide to produce apoptotic daughter cells are shown below the X-axis. Data are shown as mean ± SD. N = 3–4. Dunn’s multiple comparisons test was used to assess statistical significance, with MSpppaa (magenta), whose apoptotic daughter cell completes apoptosis over 400 min after birth, as a control. *p<0.05, **p<0.01, ***p<0.001.
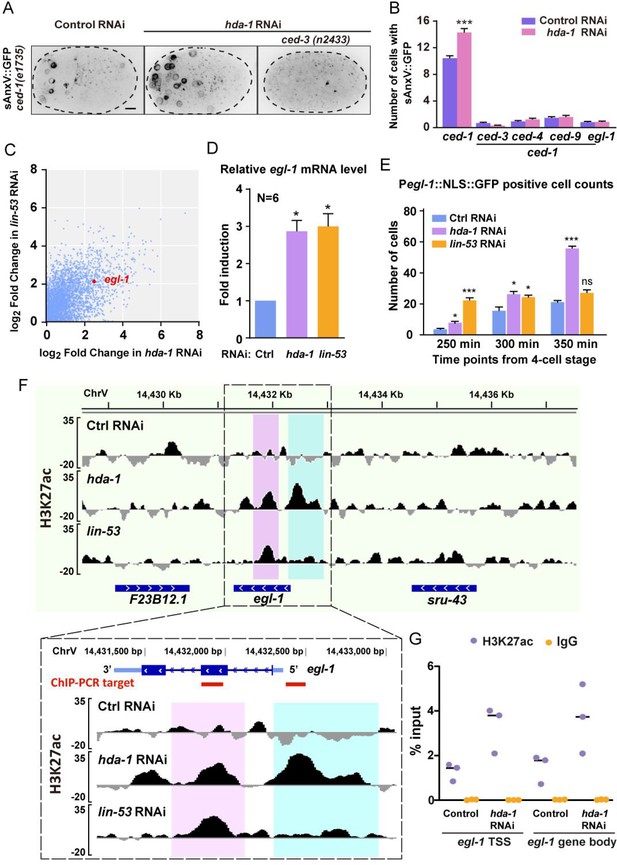
RNAi of hda-1 induces ectopic apoptosis and increases H3K27 acetylation of the egl-1 gene.
(A) Representative inverted fluorescence images show Phsp::sAnxV::GFP from ced-1(e1735) and ced-1(e1735); ced-3(n2433) embryos between late gastrulation and bean stage, treated with control RNAi or hda-1 RNAi. Scale bars, 5 μm. (B) Quantifications of cell corpse number in the embryos of indicated genotypes. Data are presented as mean ± SEM. N = 39–60. Statistical significance is determined by Student’s t-test. ***p<0.001. (C) Scatter plot of the increased gene expression in both hda-1 RNAi and lin-53 RNAi animals. The egl-1 gene is marked in red. See also Supplementary file 3. (D) Quantitative real-time PCR (RT-PCR) measurement of egl-1 mRNA levels in the control, hda-1, or lin-53 RNAi embryos. Fold induction was calculated relative to levels in control RNAi embryos. Data of six biological replicates are presented as mean ± SEM. Statistical significance is determined by the Wilcoxon test as 1 as the theoretical median. *p<0.05. (E) Quantification of the number of cells expressing the Pegl-1::NLS::GFP reporter in embryos treated with control, hda-1, or lin-53 RNAi. Data are presented as mean ± SEM. N = 8–9. Statistical significance is determined by Dunn’s multiple comparisons test. *p<0.05, ***p<0.001, ns: not significant. (F) Normalized ChIP-seq signal profiles of the H3K27 acetylation level in the control, hda-1, or lin-53 RNAi embryos at the egl-1 locus. The Y-axis shows the average sequencing coverage of bins per million reads of three biological replicates normalized to the input control sample. Ectopic H3K27ac enrichments at the egl-1 locus of hda-1 RNAi and lin-53 RNAi embryos are highlighted in magenta and cyan. (G) ChIP-qPCR analyses using H3K27ac antibody or IgG (the negative control) at selected elements of egl-1 indicated in (F) by red lines. Results are shown as the percentage of input DNA. Data of three biological replicates are presented.
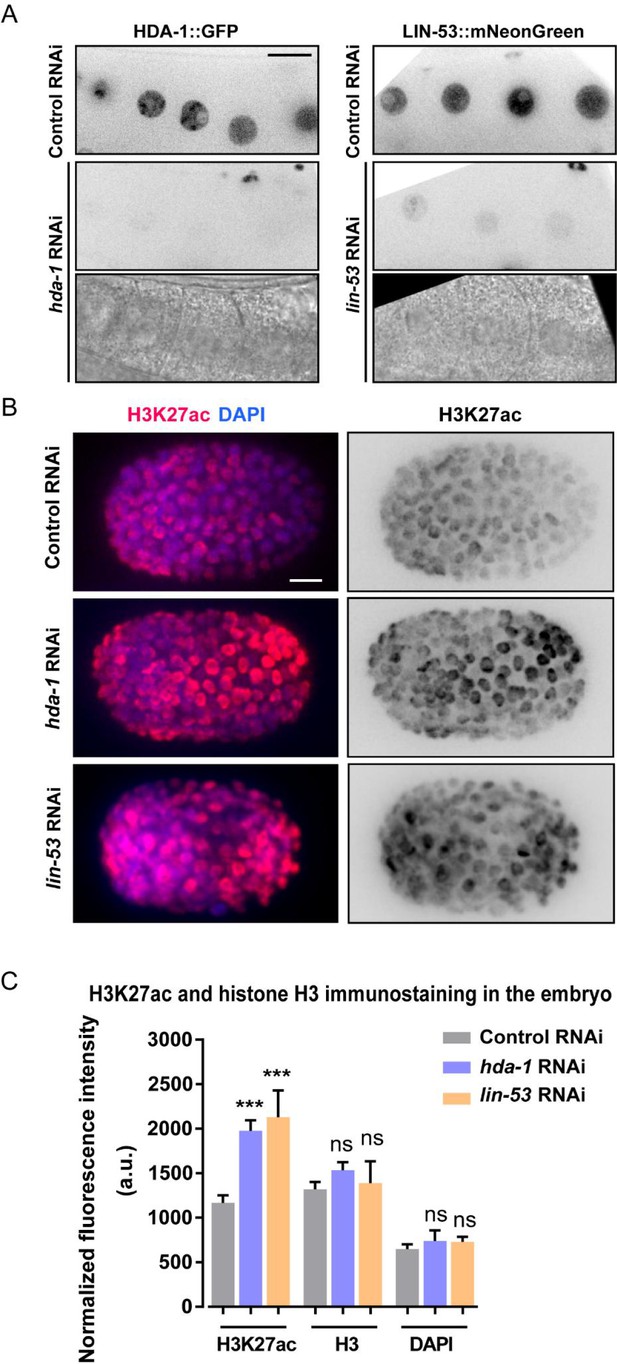
RNAi of hda-1 and lin-53 reduce fluorescence of HDA-1::GFP and LIN-53::mNeonGreen and enhance H3K27 acetylation level.
(A) Representative inverted fluorescence images (top and middle) and bright-field images (bottom) of oocytes in HDA-1::GFP (left) and LIN-53::mNeonGreen (right) KI animals treated with control, hda-1, or lin-53 RNAi. Scale bar: 5 µm. (B) Immunofluorescent images with the anti-H3K27ac antibody of control, hda-1, or lin-53 RNAi embryos around the same developmental stage. DAPI stained nuclei. Anterior of the cell is left. Scale bar: 5 µm. (C) Fluorescence intensity quantification of H3K27ac, histone H3, and DAPI from control, hda-1, or lin-53 RNAi embryos. The fluorescence intensity is arbitrary units (A.U.). Data are presented as mean ± SEM. N = 16–17. Statistical significance is determined by Student’s t-test. ***p<0.001, ns: not significant.
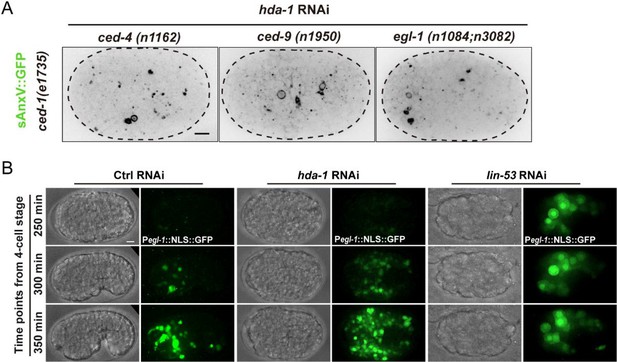
HDA-1 regulates apoptotic cell fate through the canonical apoptosis pathway.
(A) Inverted fluorescence images of Phsp::sAnxV::GFP in ced-1(e1735), ced-1(e1735); ced-4(n1162), ced-1(e1735); ced-9(n1950), or ced-1(e1735); egl-1(n1084n3082) embryos between late gastrulation stage and bean stage, treated with hda-1 RNAi. Scale bars: 5 μm. (B) Bright-field and fluorescent images of the hda-1 or lin-53 RNAi embryos that expressed the Pegl-1::NLS::GFP reporter. The anterior is to the left, and the dorsal is up. Scale bar: 5 μm.
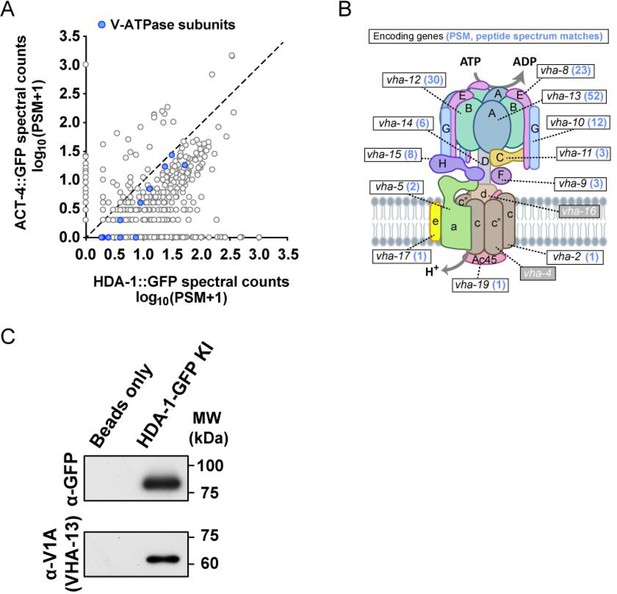
HAD-1 interacts with subunits of V-ATPase.
(A) The plot compares counts of proteins co-precipitated with HDA-1::GFP with those with the control ACT-4 (actin)::GFP. The PSM (Peptide-Spectrum Match) is the number of identified peptide spectra matched for the protein. Blue dots represent the subunits of V-ATPase. See also Supplementary file 4. (B) Schematic model of the V-ATPase complex. The known worm subunits are indicated. The mean PSM of the encoded protein from two co-IP and MS repeats is shown in blue after the gene name. (C) Western blot (WB) showing co-immunoprecipitation (co-IP) of V-ATPase V1 domain A subunit (V1A) with the HDA-1 from worm lysates. Assay was performed using three biological replicates. Three independent biological replicates of the experiment were conducted with similar results.
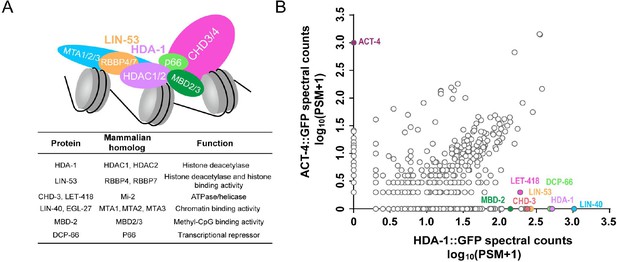
The composition of C. elegans nucleosome remodeling and deacetylase (NuRD) and NuRD subunits identified by co-IP and mass spectrometry.
(A) Upper: a schematic representation of the NuRD complex (Bracken et al., 2019; Lai and Wade, 2011). Lower: C. elegans homologs of NuRD subunits and their function. (B) Mass spectrometric analysis of proteins purified by anti-GFP agarose beads from HDA-1::GFP KI worms or transgenic worms expressing an actin protein ACT-4::GFP. The plot compares average counts of proteins from two biological replicates co-precipitated with HDA-1::GFP with protein counts co-precipitated with the control ACT-4::GFP. ACT-4::GFP affinity purification pulled down ACT-4 but none of the NuRD subunits (Y-axis), whereas HDA-1::GFP pulled down all the known NuRD components but not ACT-4 (X-axis). The number of PSM is the total number of identified peptide spectra matched for the protein. See also Supplementary file 4.
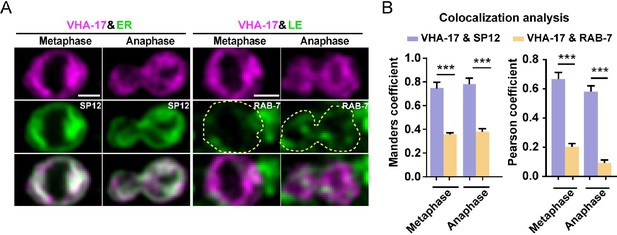
V-ATPase co-localizes with endoplasmic reticulum.
(A) Representative double-labeling images of VHA-17 and the ER marker SP12 (left) or the late endosomal marker RAB-7 (right) at metaphase and anaphase in dividing QR.a cells. Scale bar: 2 µm. (B) Quantification of VHA-17 with SP12 or RAB-7 colocalization using Manders overlap coefficient. Data are presented as mean ± SEM. N = 9–13. Statistical significance is determined by Student’s t-test. ***p<0.001.
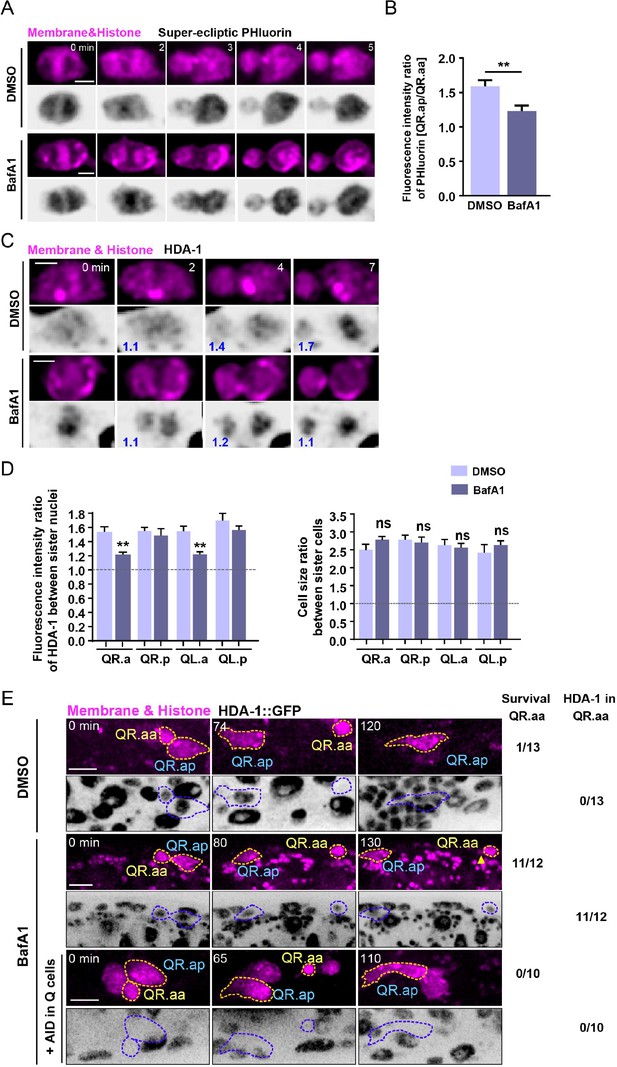
V-ATPase regulates nucleosome remodeling and deacetylase (NuRD) asymmetric segregation and cell fates.
(A) Dynamics of the cytosolic pH indicated by super-ecliptic pHluorin during QR.a division in DMSO- or BafA1-treated animals. In each panel, the top row shows mCherry-tagged plasma membrane and histone, and the bottom row shows inverted fluorescence images of super-ecliptic pHluorin. The anterior of the cell is on the left. Time 0 min is the onset of anaphase. Scale bar, 2 µm. See also Video 8. (B) The super-ecliptic pHluorin fluorescence intensity ratio between QR.ap and QR.aa in DMSO control or BafA1-treated animals. Data are presented as mean ± SEM. N = 11–15. Statistical significance is determined by Student’s t-test. **p<0.01. (C) Images of HDA-1::GFP distribution during QR.a cell division in DMSO- or BafA1-treated animals. In each panel, the top row shows mCherry-tagged plasma membrane and histone, and the bottom row shows inverted fluorescence images of HDA-1::GFP. The GFP fluorescence intensity ratios between the posterior and anterior chromatids, and between QR.ap and QR.aa nuclei are shown in blue at the lower left corner of inverted fluorescence images. Anterior of the cell is left. Scale bar: 2 µm. See also Video 9. (D) Quantification of HDA-1::GFP fluorescence intensity ratios between the large and small daughter cell nuclei and the cell size ratios between the large and small daughters. The names of mother cells are shown on the X-axis. Data are presented as mean ± SEM. N = 9–12. Statistical significance is determined by Student’s t-test. **p<0.01, ns: not significant. (E) Representative images showing fates of QR.aa after DMSO (top), BafA1 (middle), and BafA1 plus AID treatment (bottom). The Q cell plasma membrane and chromosome are labeled by mCherry. HDA-1::GFP is shown as inverted fluorescence images. Yellow arrowhead shows a short neurite-like outgrowth of QR.aa in BafA1-treated larvae. Frequencies of QR.aa survival and HDA-1 maintenance are showed on the right. Time 0 min is the birth of QR.aa. Scale bar: 5 µm.
Quantile-quantile (Q–Q) plots for the data in Figures 1F, G, 3B, 5B, D, 6B.
The D'Agostino and Pearson and Shapiro–Wilk tests were performed to test the normal distribution of the datasets at 4 and 7 min, in Figure 1F. The Shapiro–Wilk tests were performed to test the normal distribution of the data in Figures 1G, 3B, 5B, D, 6B. p-Values from Shapiro–Wilk tests are given in parentheses. For each panel, three or more experimental replicates are performed.
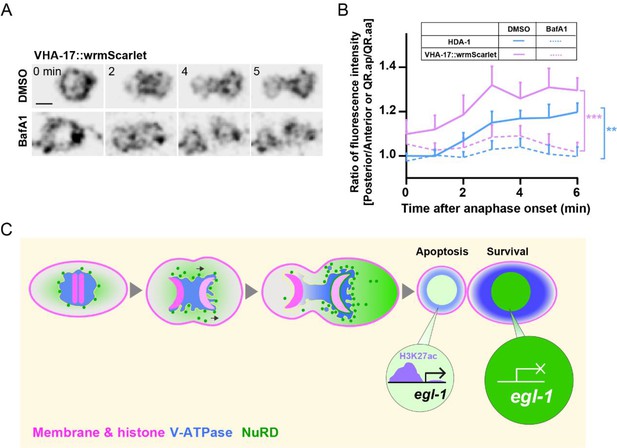
V-ATPase distribution during asymmetric cell divisions (ACDs) and a model.
(A) Dynamics of VHA-17::wrmScarlet during QR.a cell division in DMSO- or BafA1-treated animals. VHA-17::wrmScarlet fluorescence is shown as inverted fluorescence images. Time 0 min is the onset of anaphase. Scale bar: 2 µm. See also Video 10. (B) Quantification of VHA-17::wrmScarlet (magenta) and HDA-1::GFP (blue) fluorescence intensity ratios between the posterior and anterior half of QR.a or between QR.ap and QR.aa. Data are presented as mean ± SEM. N = 10–14. Student’s t-test was used to compare the intensity ratio difference of VHA-17 and HDA-1 between DMSO- and BafA1- treated cells at 6 min after anaphase. **p<0.01, ***p<0.001. (C) A proposed model. Asymmetric segregation of V-ATPase mediated the enrichment of its associated nucleosome remodeling and deacetylase (NuRD) in the large daughter cell, where high level of NuRD deacetylates egl-1 and suppressed its expression.
Videos
Dynamics of CHD-3 during QR.a division.
Fluorescence time‐lapse movies of CHD-3::GFP (green) and mCherry-labeled plasma membrane and histone (magenta) in QR.a. Frames were taken every 1 min. The display rate is three frames per second. CHD-3 was asymmetrically segregated into the future surviving QR.ap. Scale bar: 2 μm.
Dynamics of MEP-1 during QR.a division.
Fluorescence time‐lapse movies of MEP-1::GFP (green) and mCherry-labeled plasma membrane and histone (magenta) in QR.a. Frames were taken every 1 min. The display rate is three frames per second. MEP-1 was asymmetrically segregated into the future surviving QR.ap. Scale bar: 2 μm.
Dynamics of HDA-1 during QR.a division.
Fluorescence time‐lapse movies of HDA-1::GFP (KI; green) and mCherry-labeled plasma membrane and histone (magenta) in QR.a. Frames were taken every 1 min. The display rate is three frames per second. HDA-1 was asymmetrically segregated into the future surviving QR.ap. Scale bar: 2 μm.
Dynamics of LIN-53 during QR.a division.
Fluorescence time‐lapse movies of LIN-53::mNeonGreen (KI; green) and mCherry-labeled plasma membrane and histone (magenta) in QR.a. Frames were taken every 1 min. The display rate is three frames per second. LIN-53 was asymmetrically segregated into the future surviving QR.ap. Scale bar: 2 μm.
Dynamics of MYS-1 during QR.a division.
Fluorescence time‐lapse movies of MYS-1::GFP (green) and mCherry-labeled plasma membrane and histone (magenta) in QR.a. Frames were taken every 1 min. The display rate is three frames per second. Anterior, left. MYS-1 was asymmetrically segregated into the future surviving QR.ap. Scale bar: 2 μm.
Dynamics of HDA-1 during QR.a division in the pig-1 mutant.
Fluorescence time‐lapse movies of HDA-1::GFP (KI; green) and mCherry-labeled plasma membrane and histone (magenta) during QR.a division in the pig-1 mutant. Frames were taken every 1 min. The display rate is three frames per second. Anterior, left. HDA-1 was evenly segregated into two daughter cells. Scale bar: 2 μm.
Dynamics of LIN-53 during QR.a division in the pig-1 mutant.
Fluorescence time‐lapse movies of LIN-53::mNeonGreen (KI; green) and mCherry-labeled plasma membrane and histone (magenta) during QR.a division in the pig-1 mutant. Frames were taken every 1 min. The display rate is three frames per second. Anterior, left. LIN-53 was evenly segregated into two daughter cells. Scale bar, 2 μm.
Dynamics of super-ecliptic PHluorin during QR.a division.
Fluorescence time‐lapse movies of super-ecliptic PHluorin (green) and mCherry-labeled plasma membrane and histone (magenta) during QR.a division in DMSO and BafA1-treated animals. Frames were taken every 1 min. The display rate is three frames per second. Anterior, left. Scale bar, 2 μm.
BafA1 treatment disrupts HDA-1 asymmetry during QR.a division.
Fluorescence time‐lapse movies of HDA-1::GFP (KI; green) and mCherry-labeled plasma membrane and histone (magenta) during QR.a division in DMSO and BafA1-treated animals. Inverted fluorescence movie of HDA-1::GFP was shown below the merged movie. Frames were taken every 1 min. The display rate is three frames per second. Anterior, left. Scale bar: 2 μm.
Dynamics of VHA-17 during QR.a division.
Fluorescence time‐lapse movies of wrmScarlet-tagged VHA-17 (magenta) and HDA-1::GFP (KI; green) during QR.a division after DMSO or BafA1 treatments. Inverted fluorescence movie of VHA-17::wrmScarlet is shown below the merged movie. Frames were taken every 1 min. The display rate is three frames per second. Anterior, left. Scale bar: 2 μm.
Tables
C. elegans strains in this study.
| Strain name | Genotype | Method |
|---|---|---|
| N2 | Wild-type | N.A. |
| GOU4633 | cas1133[hda-1::TEV-S::gfp knock-in] V; ujIs113[Ppie-1::H2B::mCherry, Pnhr-2::HIS-24::mCherry, unc-119(+)] II | Microinjection |
| SYS1031 | sys1031[lin-53::mNeonGreen knock-in] I; ujIs113[Ppie-1::H2B::mCherry, Pnhr-2::HIS-24::mCherry, unc-119(+)] II | Microinjection |
| GOU4279 | cas1133; casIs165[Pegl‐17:: myri‐mCherry, Pegl‐17::mCherry‐TEV‐S::his‐24, unc‐76(+)] II | Genetic cross |
| GOU4277 | sys1031; casIs165 | Genetic cross |
| GOU4636 | casEX873[Phda-1::hda-1::gfp::unc-54 3’UTR, Pegl‐17:: myri‐mCherry, Pegl‐17::mCherry‐TEV‐S::his‐24] | Microinjection |
| GOU4635 | casEx874[Plin-53::lin-53::gfp::unc-54 3’UTR, Pegl‐17:: myri‐mCherry, Pegl‐17::mCherry‐TEV‐S::his‐24] | Microinjection |
| GOU4637 | casEx877[Pchd-3::chd-3::gfp::UNC-54 3’UTR, Pegl‐17:: myri‐mCherry, Pegl‐17::mCherry‐TEV‐S::his‐24] | Microinjection |
| GOU3631 | wgIs70[mep-1::TY1::EGFP::3xFLAG, unc-119(+)] III; casIs165[Pegl‐17:: myri‐mCherry, Pegl‐17::mCherry‐TEV‐S::his‐24, unc‐76(+)] II | Genetic cross |
| GOU4634 | casEX890[Pmys-1::mys-1::gfp-unc-54UTR,Pegl‐17:: myri‐mCherry, Pegl‐17::mCherry‐TEV‐S::his‐24] | Microinjection |
| CU3509 | ced-1(e1735) I; smIs76[Phsp-16.41::sAnxV::gfp] | Genetic cross |
| GOU3922 | ced-3(n2433) IV; ced-1(e1735) I; smIs76[Phsp-16.41::sAnxV::gfp] | Genetic cross |
| GOU3923 | ced-4(n1162) III; ced-1(e1735) I; smIs76 | Genetic cross |
| GOU3924 | ced-9(n1950) III; ced-1(e1735) I; smIs76 | Genetic cross |
| GOU3925 | egl-1(n1084n3082) V; ced-1(e1735) I; smIs76[Phsp-16.41::sAnxV::gfp] | Genetic cross |
| smIs89 | smIs89[Pegl-1::NLS::GFP] | Microinjection |
| GOU4285 | cas1133; casIs165; pig-1(gm344) | Genetic cross |
| GOU4281 | sys1031; casIs165; pig-1(gm344) | Genetic cross |
| GOU4287 | cas1133; casEx5309[Phsp-16.2::egl-20, Pegl‐17:: myri‐mCherry, Pegl‐17::mCherry‐TEV‐S::his‐24] | Genetic cross |
| GOU4283 | sys1031; casEx5309[Phsp-16.2::egl-20, Pegl‐17:: myri‐mCherry, Pegl‐17::mCherry‐TEV‐S::his‐24] | Genetic cross |
| GOU4638 | cas1133; him-5(e1490) V | Genetic cross |
| GOU4204 | sys1031; him-5(e1490) V | Genetic cross |
| GOU4607 | cas1589 [hda-1::GSlinker::degron::TEV-S::gfp knock-in];casIs165; casEx900[Pegl‐17:: TIR1::mRuby::unc-54 3’UTR,odr-1::gfp] | Microinjection |
| EX906 | casEx906 [Pegl-17::vha-17::wrmScarlet::unc-54 3’UTR;odr-1::gfp;Pegl-17::TIR1:unc-54 3’UTR];cas1589 | Microinjection |
| EX913 | casEx913[Pegl-17::sp12::gfp::unc-54 3’UTR;Pegl-17::vha-17::wrmScarlet::unc-54 3’UTR; odr-1::gfp] | Microinjection |
| SYB4702 | syb4702[gfp::rab-7 knock-in] | Microinjection: syb4702 was generated by Suny Biotech (http://www.sunybiotech.com/) using CRISPR-Cas9 |
| EX909 | casEx909[Pegl-17::vha-17::wrmScarlet::unc-54 3’UTR;odr-1::gfp]; syb4702 | Microinjection |
| EX910 | casEx910 [Pegl-17::super-ecliptic PHluorin::unc-54 3’UTR; odr-1::rfp]; casIs165 | Microinjection |
| EX911 | casEx910; syb4796 | Genetic cross |
Genomic targets for CRISPR.
| Gene | CRISPR‐Cas9 targets (PAM) |
|---|---|
| hda-1 knock‐in | sg1: CTTCTACGATGGTGAGCGTGAGG |
| hda-1 knock‐in | sg2: GCAGCTCAGTTTGAGTCGGAAGG |
| lin-53 knock‐in | sg1: ATTTGCGACGCGATCTTCGGAGG |
| lin-53 knock‐in | Sg2: GGAGGTTCCATCTTCAAGAGTGG |
| vha-2 knock-in | sg1: ATCATTCCGACGAAGAGTCTTGG |
| vha-2 knock-in | sg2: GACGAAGAGTCTTGGCTGTTGGG |
| rab-7 knock‐in | sg1: TTTGAGCAGCGCCTTCTTTCTGG |
| rab-7 knock‐in | sg2: AATGTCGGGAACCAGAAAGAAGG |
Plasmids and primers used in this study.
| Plasmids or PCR products | Forward primer | Reverse primer | Notes |
|---|---|---|---|
| pDD162-Peft‐3::Cas9+PU6::hda-1 sg1 | TCTACGATGGTGAGCGTGG TTTTAGAGCTAGAAATAGC | CGCTCACCATCGTAGAAG CAAGACATCTCGCAATAGGA | PCR from pDD162-Peft-3::Cas9+PU6::Empty sgRNA |
| pDD162-Peft-3::Cas9+PU6::hda-1 sg2 | AGCTCAGTTTGAGTCGGAG TTTTAGAGCTAGAAATAGC | CGACTCAAACTGAGCTGCC AAGACATCTCGCAATAGGA | PCR from pDD162-Peft-3::Cas9+PU6::Empty sgRNA |
| pPD95.77-hda-1–5’ arm::3’ arm knock-in | GTACCGGTAGAAAAAAT GAACTCAAACGGCCCGTT | GGAATTCTACGAATGCGAA TAAACCCTTGCGGCTT | The 5’ arm::hda-1::3’ arm sequences were amplified from N2 and cloned into pPD95.77 via In‐Fusion Advantage PCR Cloning Kit |
| pPD95.77-hda-1–5’ arm::TEV-S::gfp::3’ arm knock-in | GAACTATACAAATAGAACA CTAAAATGTGCCGCCG | CCGATCCCCCGGGCACT CTGTCTTCTGACGCTTTT | The TEV-S-gfp was cloned into pPD95.77-hda-1–5’ arm::3' arm knock-in via In-Fusion Advantage PCR Cloning Kit |
| pPD95.77-hda-1::TEV-S::gfp knock-in repair template | GATGGTGAGCGTGAAGGAGAT | CTTCACGCTCACCATCGTAG | PCR on pPD95.77-hda-1–5’ arm::TEV-S::gfp::3’arm knock-in plasmid to synonymously mutate the PAM sequence of sg2 |
| Pegl-17:: myri-mCherry | CTTCCGTTCTATGGAACACTC | GAATCATCGTTCA CTTTTCACGG | Pegl-17 promoter was amplified from N2 genomic DNA and inserted into the pDONR P4-P1R-mCherry plasmid via In-Fusion Advantage PCR Cloning Kit |
| Pegl-17::mCherry ::TEV-S::his-24 | CTTCCGTTCTATGGAACACTC | GAAGACGTTGAACG TCAAATTATC | Pegl-17 promoter was amplified from N2 genomic DNA and inserted into the pDONR P4-P1R-mCherry::TEV-S::his-24 plasmid via In-Fusion Advantage PCR Cloning Kit |
| linker::gfp::unc-54_3'UTR | AGACCCAAGCTTGGTACCA TGAGTAAAGGAGAAGAACTTTTCAC | AAGGGCCCGTACGGCC GACTAGTAGG | PCR from the plasmid pPD95.77 and then used as SOEing PCR template |
| Phda-1::hda-1 | CCAACTTCGACCTCACCCTC | GGTACCAAGCTTGGGTCTCTCT GTCTTCTGACGCTTTT | PCR from N2 genome and was then used as SOEing PCR template |
| Plin-53::lin-53 | AGCAAATGTTGCAGGGCTGTG | GGTACCAAGCTTGGGTCTCT GTTGTCTCTCTACCACAT | PCR from N2 genome and was then used as SOEing PCR template |
| Pchd-3::chd-3 | CACCTGTCCTTCGTGCCTATC | GGTACCAAGCTTGGGTCTAT ATCTCGGATAGGACGAACC | PCR from N2 genome and was then used as SOEing PCR template |
| Pmys-1::mys-1 | GCTCGTTATCAAGAAGGTCTCC | ACCAAGCTTGGGTCTGAA CATGATCTGCGCCTGAA | PCR from N2 genome and was then used as SOEing PCR template |
| Phda-1::hda-1::linker::gfp::unc-54_3'UTR | ACCAGTGCTCGACTTCGTGATG | GGAAACAGTTATGTTT GGTATATTGGG | SOEing PCR |
| Plin-53::lin-53::linker::gfp::unc-54_3'UTR | AGTCGGTCTTTGCGCTCAAC | GGAAACAGTTATGTTT GGTATATTGGG | SOEing PCR |
| Pchd-3::chd-3::linker::gfp::unc-54_3'UTR | GATCGTTGGTTAGG TCTCTCATGG | GGAAACAGTTATGTTT GGTATATTGGG | SOEing PCR |
| Pmys-1::mys-1::linker::gfp::unc-54_3'UTR | ATAAGAGCAAGAGT CAAGGCAGTC | GGAAACAGTTATGTTT GGTATATTGGG | SOEing PCR |
| egl-1 | GGCTACGAGATCGGCTCCAA | GAAGCATGGGCCGAGTAGGA | RT-qPCR |
| cdc-42 | GGAATGCTCGAGAAACTGGC | CAGTCCCTTCTGCGTCAAC | RT-qPCR |
| egl-1 TSS region | TAATCATCCTCATCAAGCCTGC | CACAGCTTCTCATTGCACGC | ChIP-qPCR |
| egl-1 gene body region | CTCTTCGGATCTTCTACCAATGTC | GAGTCGTCGGCAAATTGAGA | ChIP-qPCR |
| pPD95.77- Pegl-17::TIR1::mRuby::unc-54 3’UTR | ATGCAAAAGAGAATCGCCTTGT | AACAGTTATGTTTGGT ATATTGGGAATG | TIR1::mRuby::unc-54 3’UTR fragment was amplified from strain CA1210:IE28[dhc1::degron::gfp];ieSo57[Peft-3::TIR1::mRuby::unc-54 3'UTR.unc-119(+)]II |
| pPD95.77- Pegl-17::vha-17::wrmScarlet::unc-54 3’UTR | cccgaaatgtgagc tATGGGTATT CTCATTCCA CTCGTC | GCTACCACTTCCAGCGTTG TTGATTACGTTTGGTGCG | vha-17 genomic fragment was PCR from N2 genome and inserted into the pPD95.77- Pegl-17::wrmScarlet::unc-54 3’UTR plasmid via In-Fusion Advantage PCR Cloning Kit |
| pPD95.77-Pegl-17::sp12::gfp::unc-54 3’UTR | gcccgaaatgtgag ctATGGACG GAATGATTG CAATGC | GCTACCACTTCCAGCTTTC GTCTTCTTTGTCTCCTTTTTC | sp12 genomic fragment was PCR from N2 genome and inserted into the pPD95.77- Pegl-17::wrmScarlet::unc-54 3’UTR plasmid via In-Fusion Advantage PCR Cloning Kit |
| pPD95.77-Pegl-17::super-ecliptic PHluorin::unc-54 3’UTR | cccgaaatgtgagc tATGAGTAA AGGAGAAG AACTTTTCA | GAAGAGTAATTGGACCTATTTG TATAGTTCATCCATGCCA | Super-ecliptic PHluorin fragment was synthesized and inserted into the pPD95.77- Pegl-17::wrmScarlet::unc-54 3’UTR plasmid via In-Fusion Advantage PCR Cloning Kit |
Additional files
-
Supplementary file 1
Single-cell SPLiT-seq expression matrix and genes that were not detected from the Pegl-1-NLS-gfp-positive cells.
- https://cdn.elifesciences.org/articles/89032/elife-89032-supp1-v1.xlsx
-
Supplementary file 2
The fluorescence intensity ratio of each cell pair and normalized fluorescence intensity of each cell in embryonic lineage tracing assay.
- https://cdn.elifesciences.org/articles/89032/elife-89032-supp2-v1.xlsx
-
Supplementary file 3
Differential expression of RNA-seq under control, hda-1, or lin-53 RNAi.
- https://cdn.elifesciences.org/articles/89032/elife-89032-supp3-v1.xlsx
-
Supplementary file 4
Anti-GFP IP-MS results of HDA-1::GFP KI worms and ACT-4::GFP transgenic worms.
- https://cdn.elifesciences.org/articles/89032/elife-89032-supp4-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/89032/elife-89032-mdarchecklist1-v1.docx





