Quorum-sensing agr system of Staphylococcus aureus primes gene expression for protection from lethal oxidative stress
Figures
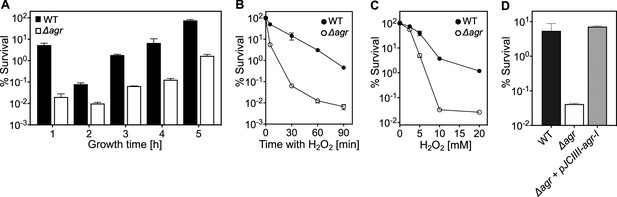
agr protects from killing by H2O2 throughout the growth cycle.
(A) Effect of culture growth phase. Overnight cultures of S. aureus LAC wild-type (WT, BS819) or Δagr (BS1348) were diluted (OD600∼0.05) into fresh TSB medium and grown with shaking from early exponential (1 h, OD600∼0.15) through late log (5 h, OD600∼4) phase. At the indicated times, early (undiluted) and late exponential phase cultures (diluted into fresh Tryptic Soy Broth (TSB) medium to OD600∼0.15) were treated with H2O2 (20 mM). After 60 min, aliquots were removed, serially diluted, and plated for determination of viable counts. Percent survival was calculated relative to a sample taken at the time of H2O2 addition. (B) Kinetics of killing by H2O2. Wild-type and Δagr mutant strains were grown to early exponential (OD600∼0.15) and treated with 20 mM H2O2 for the times indicated, and percent survival was determined by plating. (C) Effect of H2O2 concentration on survival. Cultures prepared as in panel B were treated with the indicated peroxide concentrations for 60 min prior to plating and determination of percent survival. (D) Complementation of agr deletion mutation. Cultures of wild-type (WT) cells (BS819), Δagr mutant (BS1348), and complemented Δagr mutant carrying a chromosomally integrated wild-type operon (pJC1111-agrI) were treated with 20 mM H2O2 for 60 min followed by plating to determine percent survival. Data represent the means ± SD. from biological replicates (n=3).
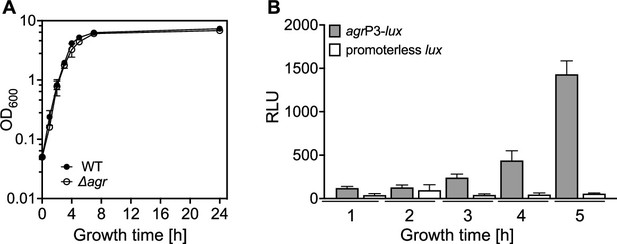
Correlation of growth phase and agr expression.
(A) Growth curves. Overnight cultures of S. aureus LAC wild-type (WT, BS819) or Δagr mutant (BS1348) were diluted (OD600∼0.05) in fresh TSB medium and growth was monitored by measuring the optical density at 600 nm (OD600). (B) Tests of agrP3 promoter activity. S. aureus LAC wild-type (WT, BS819) containing agrP3-lux (SaPI1 attC::agrP3-lux; strain BS1222) or control containing a promoterless lux gene within the attC site (SaPI1 attC::pGYLux, strain BS999) grown as in (A) for the indicated times. agrP3 activity (relative luminescence units [RLU]) was assayed at the indicated times (see Materials and methods). Data represent the means ± SD. from biological replicates (n=3).
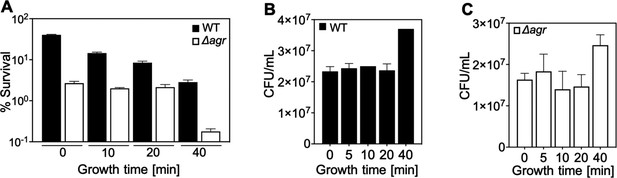
Correlation of lag-time and agr-mediated protection from H2O2-mediated killing.
Overnight cultures of S. aureus LAC wild-type (WT, BS819) and Δagr mutant (BS1348), grown for the indicated times following dilution to fresh medium, were treated with H2O2 (20 mM for 60 min) (Figure 1A). Data represent the means ± SD. from biological replicates (n=3). Survival of Δagr mutant cells was unchanged up to the 40 min time point, and then it dropped sharply. The sharp drop coincided with the time to first division (i.e. the lag time), as evidenced by an increase in colony-forming units (CFUs) at the 40 min time point in the absence of treatment (B and C). In contrast to results with the Δagr strain, survival of the wild-type strain gradually decreased throughout the experiment (A). Increased lag-time is associated with tolerance to lethal stress owing to a delay in growth when switched to a new environment (Fridman et al., 2014). Thus, our observations suggest that a subpopulation of Δagr mutant cells remains longer in a dormant state, decreasing the lethality of H2O2. The differential effect of the lag time on the wild-type and Δagr mutant cultures was absent during exponential growth (40 min). These results suggest that agr contributes to at least two forms of protection from H2O2-mediated killing: tolerance by a transient lag state and tolerance during growth phase. To focus on the latter form, assays involving cultures after overnight growth were grown for ~65 min (OD600∼0.15).
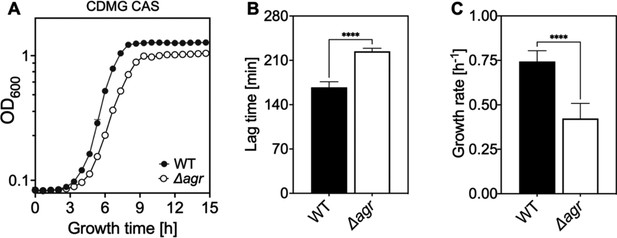
Extended lag phase and decreased growth rate and yield of an Δagr mutant.
(A) Growth curves. S. aureus LAC wild-type (WT, BS819) and Δagr mutant (BS1348) cultures were grown in chemically-defined medium supplemented with 0.5% Casamino acids and 14 mM glucose (CDMG CAS) for the indicated times following 1000-fold dilution of overnight cultures grown in TSB. Growth of diluted cultures was monitored for 15 hr every 40 min by measuring the OD600 using an Agilent LogPhase 600 Microbiology Reader (Santa Clara, CA). (B) Lag times. Data in panel A were used to determine lag times by extrapolation of the linear portion of the growth curve. Growth rates (µ, h−1) calculated from five biological replicates are displayed in panel (B). Data are mean ± SD. Statistical significance was calculated with Student’s two-tailed t-test (****p≤0.0001).
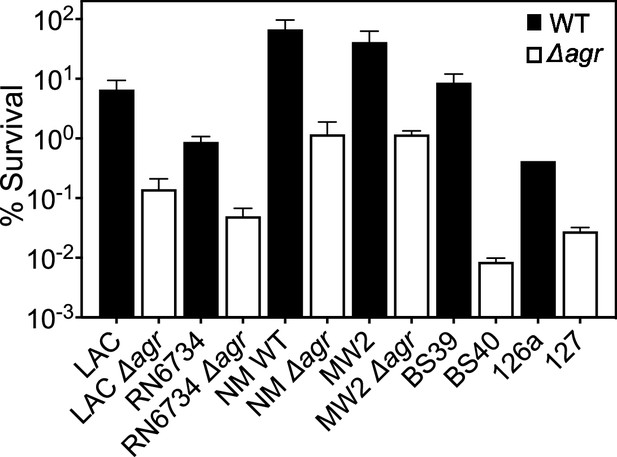
Agr-mediated protection from H2O2-mediated killing among diverse S. aureus strains.
Laboratory strains LAC, RN6734, Newman (NM, BS12), MW2 (BS450), and clinical isolates BS39 and 126 a with agr deficient mutant derivatives were compared for survival following treatment with 20 mM of H2O2 for 60 min. In this experiment, overnight cultures were diluted in Tryptic Soy Broth (TSB) and grown to early log phase (OD600∼0.15). Percent survival was determined relative to samples taken at the time of peroxide treatment. Some mutants were created by transduction of marker-disrupted alleles (LAC, RN6734, Newman, MW2) while others were naturally occurring (BS40, 127) (see Table 1). Data represent the mean ± SD. from biological replicates (n=3). The data show that peroxide lethality varies among strains, but in each case, deletion of agr increases killing.
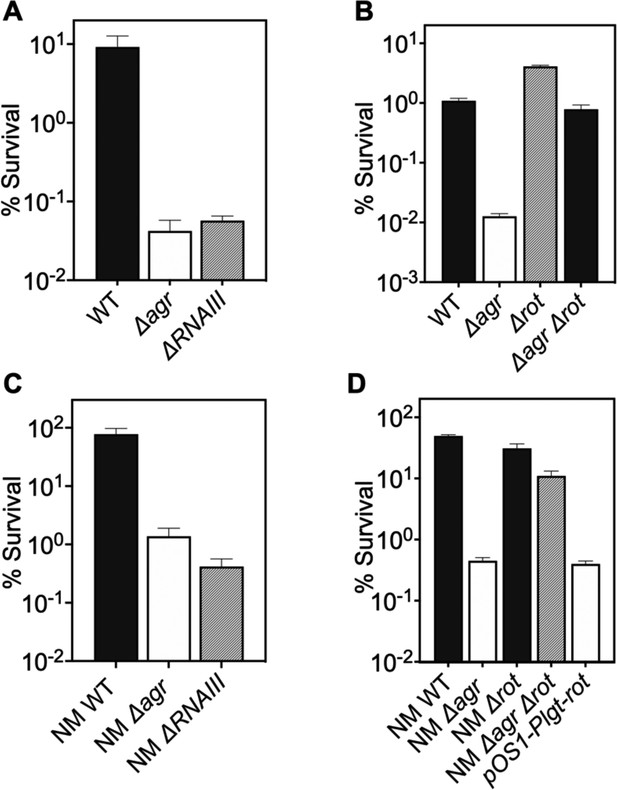
Involvement of RNAIII and rot-dependent pathways in agr-mediated protection from H2O2-mediated killing.
Cultures were grown for 1 hr following dilution from overnight cultures to early log phase (OD600∼0.15) and then treated with 20 mM H2O2 for 60 min before determination of percent survival by plating and enumeration of colonies. (A) Wild-type LAC (WT, BS819), Δagr mutant (BS1348), and ΔrnaIII mutant (GAW183). (B) Δrot and Δagr Δrot double mutant (BS1302). (C) Wild-type (WT) strain Newman (NM, BS12), Δagr mutant (BS13), and ΔRNAIII mutant (BS669). (D) Overexpression of rot. Rot was expressed from a plasmid-borne wild-type rot (pOS1-Plgt-rot, strain VJT14.28). Data represent the mean ± SD. from biological replicates (n=3).
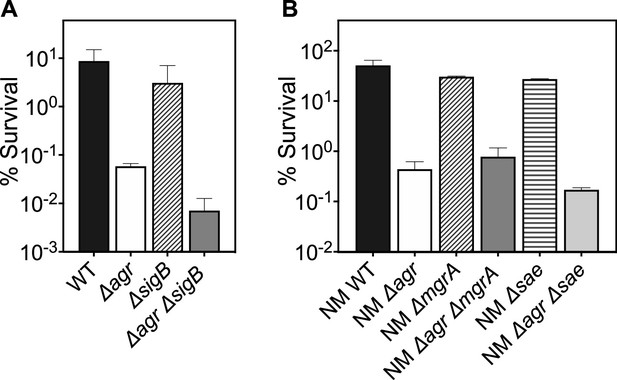
Deficiency of downstream global regulators does not differentially affect agr-mediated protection from H2O2-mediated cell death.
The effect of (A) sigB, (B) mgrA, and sae on survival in the presence or absence of agr during treatment with H2O2 was measured. Cells were grown to early log phase (OD600∼0.15) and treated with 20 mM of H2O2 for 60 min. Data represent the mean ± SD. from biological replicates (n=3). Bacterial strains were BS819 (LAC) and BS12 (Newman, NM) for wild-type (WT) and BS1348 (LAC) and BS13 (NM) for the agr, and BS1435-36, BS1280, BS1282, and BS1246, BS1518 for sigB, sae and mgrA mutants, respectively. The genes tested were either part of known two-component systems or SarA protein-family regulatory circuits involved in virulence gene expression. They are all downstream/epistatic to agr-RNAIII (reviewed in Bronesky et al., 2016). Mutations in sigB, mgrA, and sae showed little or no effect with respect to the protective agr-mediated phenotype. These results support the idea that rot is the primary regulator pathway that protects the wild-type from H2O2-mediated killing.
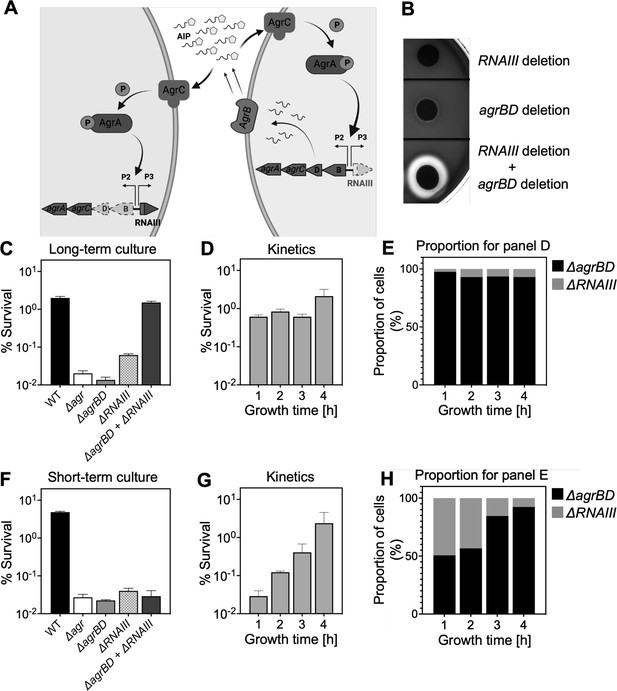
Agr-mediated protection from H2O2 stress is uncoupled from agr activation kinetics.
(A) Assay design. An ΔagrBD deletion mutant (GAW130) was complemented in trans by the autoinducing product (AIP) of AgrBD in an ΔrnaIII (GAW183) mutant that produces AIP endogenously; AgrC activation in the ΔagrBD strain leads to downstream activation of RNAIII. The agrBD strain, engineered in-frame to avoid polar effects on downstream genes agrC and agrA, senses but does not produce an autoinducer. The ΔrnaIII mutant, constructed by replacement of rnaIII with a cadmium resistance cassette (rnaIII::cadA), produces autoinducer but lacks RNAIII, the effector molecule of agr-mediated phenotypes with respect to H2O2. (B) Trans-activation demonstrated by hemolysin activity on sheep blood agar plates. Bottom of figure shows zone of clearing (hemolysin activity) after mixing 108 ΔagrBD CFU with an equal number of ΔrnaIII. Zone of clearance is a consequence of AgrC receptor activation in trans by AIP produced by the ΔrnaIII mutant. (C) Absence of trans-activation with short-term culture. The wild-type strain RN6734 (WT, BS435), ΔrnaIII (GAW183), ΔagrBD (GAW130), and ΔrnaIII and ΔagrBD mutants were mixed 1:1 immediately before growth from overnight culture. Overnight cultures were diluted (OD600∼0.05) into fresh Tryptic Soy Broth (TSB) medium, mixed, and grown to early log phase (OD600∼0.15) when they were treated with 20 mM H2O2 for 60 min and assayed for percent survival by plating. (D) Kinetics of killing by H2O2. Survival assays employing ΔrnaIII and ΔagrBD mixtures, performed as in panel C, but grown from early exponential (1 hr, OD600∼0.15) through late log (5 hr, OD600∼4) phase in TSB. Cultures were treated with H2O2 (20 mM for 1 hr) at the indicated time points. (E) Proportion of mixed population for panel D represented by each mutant after incubation. The ΔagrBD mutant contained an erythromycin-resistance marker to distinguish the strains following plating of serial dilutions on TS agar with or without erythromycin (5 μg/). Data represent the mean ± SD. from biological replicates (n=3). (F) Trans-activation during long-term culture. The wild-type strain RN6734 (WT, BS435), ΔrnaIII (strain GAW183), ΔagrBD (strain GAW130), and ΔrnaIII and ΔagrBD mutants mixed 1:1 prior to overnight culture. Survival assays employing ΔrnaIII and ΔagrBD mixtures, performed as in panel C. (G) Kinetics of killing by H2O2. Survival assays employing ΔrnaIII and ΔagrBD mixtures, performed as in panel D. Cultures were treated with H2O2 (20 mM for 1 hr) at the indicated time points. (H) Proportion of mixed population for panel G represented by each mutant after incubation, performed as in panel E. Data represent the mean ± SD. from biological replicates (n=3).
© 2024, BioRender Inc. Panel A was created with BioRender.com and is published under a CC BY-NC-ND 4.0. Further reproductions must adhere to the terms of this license
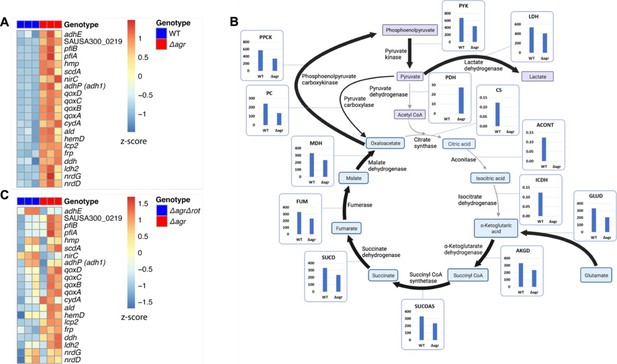
Association of agr deficiency with increased expression of respiration and fermentation genes during aerobic growth.
(A) Relative expression of respiration and fermentation genes. RNA-seq comparison of S. aureus LAC wild-type (WT, BS819) and Δagr mutant (BS1348) grown to late exponential phase (OD600~4.0). Shown are significantly up-regulated genes in the Δagr mutant (normalized expression values are at least twofold higher than in the wild-type). Heatmap colors indicate expression z-scores. RNA-seq data are from three independent cultures. See Supplementary file 1 for supporting information. (B) Schematic representation of agr-induced changes in metabolic flux, inferred from transcriptomic data (Supplementary file 1) by SPOT (Simplified Pearson correlation with Transcriptomic data). Metabolic intermediates and enzymes involved in catalyzing reactions are shown. The magnitude of the flux (units per 100 units of glucose uptake flux) is denoted by arrowhead thickness. Boxed charts indicate relative flux activity levels in wild-type versus Δagr strains. Enzyme names are linked to abbreviations in boxed charts (e.g. lactate dehydrogenase, LDH). See Supplementary file 2 for supporting information. (C) RNA-seq comparison of an Δagr Δrot double mutant (BS1302) with its parental Δagr strain (BS1348). Heatmap colors indicate expression z-scores. Sample preparation and figure labeling as for A. See Supplementary file 3 for supporting information.
© 2024, BioRender Inc. Panel B was created with BioRender.com and is published under a CC BY-NC-ND 4.0. Further reproductions must adhere to the terms of this license
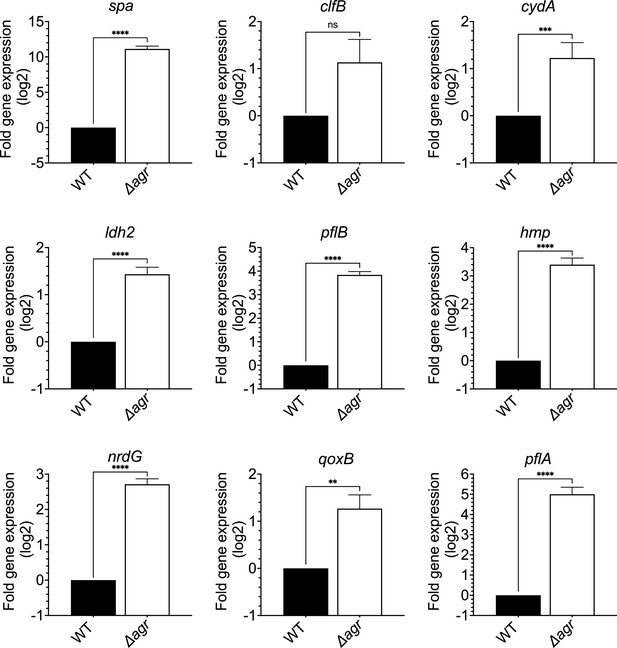
Induction of expression of selected fermentive/anaerobic genes stimulated by deletion of agr.
Total cellular RNA was extracted from late exponential phase cultures (OD600∼4.0) of wild-type (WT, BS819) or Δagr mutant (BS1348), followed by reverse transcription and PCR amplification of the indicated genes from Figure 5A, using rpoB as an internal standard. mRNA levels were normalized to those of each gene with an untreated wild-type control. Data represent the mean ± SEM of three independent experiments. Student’s t-test was used to determine statistical differences between samples (**p<0.01; ***p<0.001; ****p<0.0001). In each case the agr deletion increased expression, indicating elevated metabolism.

Association of agr deficiency with a metabolic flux shift toward fermentive metabolism during aerobic growth.
(A) Intracellular ATP levels. Comparison of S. aureus LAC wild-type (WT, BS819) and Δagr mutant (BS1348) strains for ATP expressed as µg/108 cells after growth of cultures in Tryptic Soy Broth (TSB) medium to late-exponential phase (OD600~4.0). (B) Extracellular acetate levels. Samples were taken after 1, 2, 3, 4, and 24 hr of growth in TSB medium; strains were wild-type (WT, BS819) and Δagr mutant (BS1348). (C–D) Extracellular lactate and acetate levels during low oxygen culture. S. aureus LAC wild-type (WT, BS819) and Δagr mutant (BS1348) were grown in TSB medium with suboptimal aeration to late-exponential phase (4 hr, OD600~4.0). (E–F) Oxygen consumption. Strains LAC wild-type (WT, BS819) and Δagr mutant (BS1348) were compared using Seahorse XFp analyzer (F), and the rate of oxygen consumption (E) was determined from the linear portion of the consumption curve. Representative experiments from at least three independent assays are shown. (G–H) NAD+ and NADH levels. Colorimetric assay of NAD+ (G) and NADH levels (H) for S. aureus wild-type (WT, BS819) and Δagr mutant (BS1348) after growth of cultures to late-exponential phase (OD600~4.0). (I) NAD+/NADH ratio. For all panels, data points are the mean value ± SD (n=3). *p<0.05; ****p<0.0001, by Student’s two-tailed t-test. Seahorse statistical significances are compared to TSB medium.
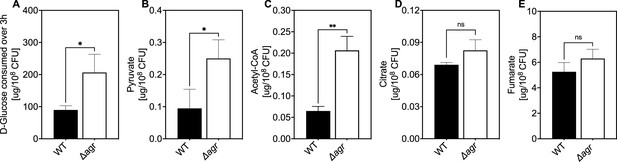
Association of agr deficiency with glucose consumption and intracellular levels of pyruvate, acetyl-CoA, and TCA-cycle metabolites.
(A) Extracellular glucose levels. D-glucose levels per expressed as expressed as µg/108 cells of S. aureus LAC wild-type (WT, BS819) or Δagr mutant (BS1348) after growth of cultures in Tryptic Soy Broth (TSB) medium to exponential phase (3 hr). (B–E) Intracellular levels of pyruvate, acetyl-CoA, and TCA-cycle metabolites citrate and fumarate. Levels of the indicated metabolite expressed as expressed as µg/108 cells of S. aureus LAC wild-type (WT, BS819) or Δagr mutant (BS1348) after growth in TSB medium to late-exponential phase (OD600~4.0). For all panels, data points are the mean value ± SD (n=3). *p<0.05; ****p<0.0001, by Student’s two-tailed t-test.
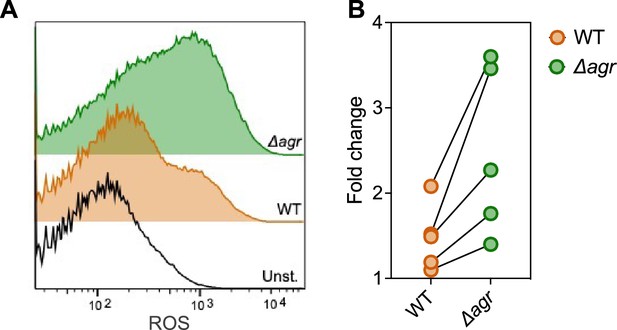
Increase in reactive oxygen species (ROS) levels associated with Δagr deficiency.
Flow cytometry measurements. S. aureus LAC wild-type (WT, BS819) and Δagr mutant (BS1348) were grown overnight, diluted, cultured in Tryptic Soy Broth (TSB) medium for 1 hr, and treated with carboxy-H2DCFDA (10 µM) for 5 min. Relative cell number is on the vertical axis. Unst. indicates samples containing LAC wild-type cells not treated with carboxy-H2DCFDA. (B) Five replicate experiments gave similar results (‘fold change’ indicates the mean wild-type or Δagr ROS level divided by the mean autofluorescence background signal; lines connect results in replicate experiments).
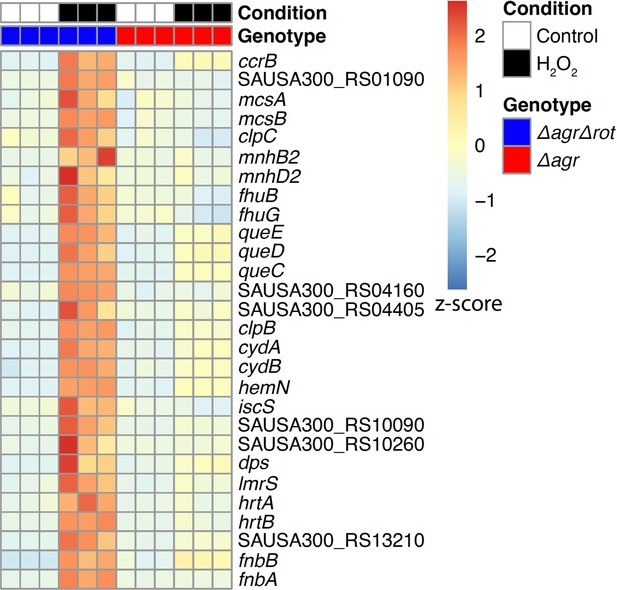
Rot-mediated up-regulation of H2O2-stimulated genes relative to those in an agr mutant.
Genes shown are those up-regulated in a Δagr Δrot double mutant (BS1302) relative to that observed with the Δagr strain (BS1348). H2O2 treatment was for 30 min. Peroxide concentrations for Δagr (2.5 mM H2O2) and Δagr Δrot (10 mM H2O2) were determined to achieve ~50% cell survival [see Methods and Figure 7—figure supplement 1]. RNA-seq data are from three independent cultures. Heatmap colors indicate expression z-scores. See Supplementary file 3 for supporting information.
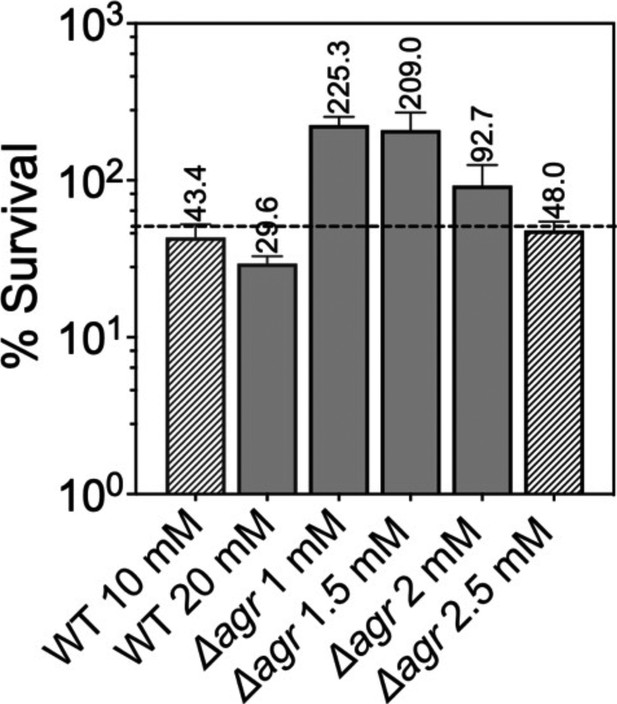
Normalization of the lethal concentration of H2O2 with wild-type and Δagr strains.
Overnight cultures were diluted into fresh Tryptic Soy Broth (TSB) medium and grown to early log phase (OD600=0.15 to achieve sufficient colony forming units (CFU) for RNA-seq). These cultures were treated with the indicated concentrations of H2O2 for 30 min prior to measurement of survival by plating. Data represent the mean ± SD. from biological replicates (n=3). Bacterial strains were BS819 for WT and BS1348 for the agr mutant. To focus RNA-seq analysis on lethal rather than cell death responses, we sought to reduce H2O2 concentrations and thereby lethality to achieve ~50% (dotted line) cell survival, normalized to wild-type and Δagr mutant strains. Survival of the Δagr mutant with H2O2 for 30 min at a concentration of 2.5 mM closely approximated 50% survival of the wild-type with 10 mM H2O2, providing a basis for choice of concentrations and treatment time for RNA-seq analysis.
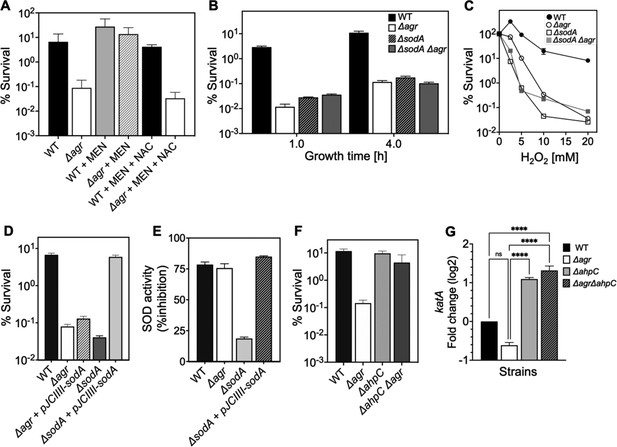
Involvement of endogenous reactive oxygen species (ROS) in agr-mediated protection from lethal H2O2 stress.
(A) Protective effect of menadione on survival. S. aureus LAC wild type (BS819) and Δagr mutant (BS1348) cultures were grown to late exponential phase (4 hr after dilution of overnight cultures), exposed to 80 μM menadione (MD) with or without 4 mM N-acetyl cysteine (NAC) for 30 min prior to treatment with H2O2 (20 mM for 1 hr) and measurement of survival. (B) Effect of sodA deletion on survival. Cultures of wild-type (BS819), Δagr (BS1348), a sodA::tetM (BS1422), and sodA::tetM-agr double mutant (BS1423) were grown to early (1 hr after dilution, OD600~0.15) or late log (4 hr after dilution, OD600~4.0) prior to treatment with 20 mM H2O2 for 60 min. (C) Effect of H2O2 concentration on survival. Late log (4 hr, OD600~4.0) cultures of the wild-type and Δagr mutant strains were treated with indicated concentrations of H2O2 for 60 min. (D) Complementation of sodA deletion mutation. A plasmid-borne wild-type sodA gene was expressed under control of the sarA constitutive promoter (pJC1111-sodA) in late log-phase (4 hr, OD600~4.0) cells treated with 20 mM H2O2 for 60 min. (E) SodA activity. Wild-type or the indicated mutants were grown to late-exponential phase (OD600~4.0); Sod activity was measured as in Methods. (F) Effect of ahpC deletion on survival. Late log-phase cultures of wild-type (BS819), Δagr (BS1348), ahpC::bursa (BS1486), and ΔahpC::bursa-agr double-mutant (BS1487) cells were treated with 20 mM H2O2 for 60 min. (G) Effect of ahpC deletion on expression of katA in the indicated mutants. Total cellular RNA was extracted from late exponential-phase cultures (OD600~4.0), followed by reverse transcription and PCR amplification of the indicated genes, using rpoB as an internal standard. mRNA levels were normalized to those of each gene to wild-type control. Data represent the mean ± SD. from (n=3) biological replicates. One-way ANOVA was used to determine statistical differences between samples (****p<0.0001).
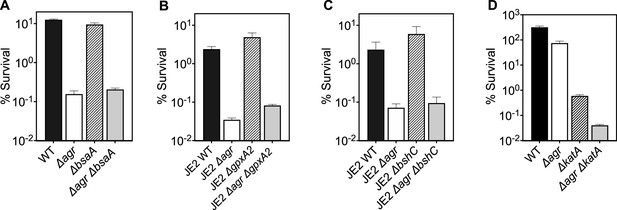
Deficiency in reactive oxygen species (ROS) detoxification genes katA, bsaA1/gpxA1, bsaA2/gpxA2, and bacilliothiol (BSH) have no effect on agr-mediated protection from H2O2-mediated cell death.
Effect of (A) bsaA (B) gpxA2, (C) bshC and (D) katA on survival during treatment with H2O2. Cells were grown to early log phase (OD6000.15) and treated with 20 mM of H2O2 for 60 min for A-C or with 2 mM of H2O2 for 60 min for D. Data represent the mean ± SD. from biological replicates (n=3). Bacterial strains were BS819 (LAC) and BS867 (LAC) for WT, and BS1348 (LAC), BS1010 (JE2), BS1490-91, BS1522-23, BS1527-28 and BS1488-89 for the agr, bsaA, gpxA2, bshC, and katA mutants, respectively. Our data with superoxide dismutases (sodA) and the peroxiredoxin ahpC (Figure 7) suggest that homeostatic detoxification pathways contribute to agr-mediated phenotypes with respect to lethal H2O2 stress. Mutations in additional genes involved in H2O2 detoxification that included catalase, (katA), two thiol-dependent peroxidases (gpxA1 and gpxA2), and the low-molecular-weight thiol bacillithiol (bshC) showed no differential effect with respect to agr-mediated phenotypes. Notably, gpxA1, which is also known as bsaA1, was essential for the oxidation-sensing ability of AgrA to confer resistance to H2O2-mediated growth inhibition (Sun et al., 2012). The ΔkatA mutation was hyperlethal with the wild-type and Δagr mutant, even when otherwise sub-inhibitory concentrations of H2O2 were used. Collectively, the data support the idea that agr-mediated phenotypes are detoxification pathway-specific.
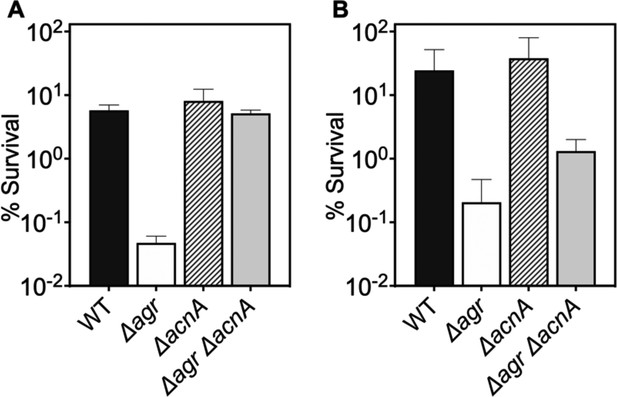
Deficiency in TCA cycle gene acnA reverses the effect of an agr deficiency with respect to subsequent challenges with H2O2.
(A–B) Effect of acnA on survival during treatment with H2O2. Cells were grown to (A) late (OD600∼4.0) or (B) early (OD600∼0.15) log phase and treated with 20 mM of H2O2 for 60 min. Bacterial strains were S. aureus LAC wild-type (WT, BS819), Δagr mutant (BS1348), acnA::bursa (BS1744), and acnA::bursa-Δagr double mutant (BS1745). Data represent the mean ± SD. from biological replicates (n=3).
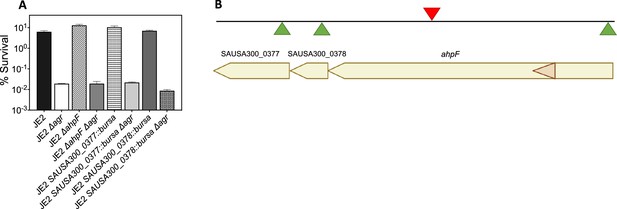
Effects of transposon insertion in ahpC unexplained by polarity of transposon insertion.
(A) Cultures of S. aureus wild-type, Δagr mutant, and various double mutants were treated with H2O2 (20 mM for 60 min) prior to measurement of survival. For strain descriptions, see Table 1. (B) ahpC locus map showing the three ORFs located downstream of ahpC. The location of four Bursa aurealis insertions (NE911, NE1571, NE537, NE725), obtained from The Nebraska Transposon Mutant Library (NTML) Fey et al., 2013 used in this study are indicated by triangles. Green triangles, plus-strand insertion; red triangle, minus-strand insertion. Data represent the mean ± SD. from biological replicates (n=3). Bacterial strains were BS435 for WT and BS1010, BS1494, BS1504, BS1495, BS1501, BS1496 and BS1506 for the agr, ahpF, SAUSA300_0377, and SAUSA300_0378 mutants, respectively. Since the Bursa aurealis (bursa) transposon insertion in ahpC was upstream of several open reading frames (ORFs) in the ahpC-F operon, polarity could complicate interpretation of the results. We, therefore, analyzed the effects of the three bursa mutants in strain JE2 downstream genes: ahpF, SAUSA300_0378, SAUSA300_0377. We found that polar effects on downstream elements could not explain the properties of ahpC::bursa. Thus, ahpC::bursa could provide insights into the role of ahpC in agr-mediated phenotypes.
© 2024, BioRender Inc. Panel B was created with BioRender.com and is published under a CC BY-NC-ND 4.0. Further reproductions must adhere to the terms of this license
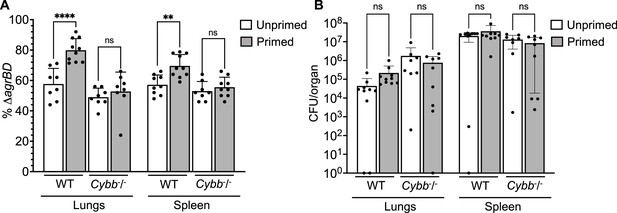
Survival advantage of agr priming of S.aureus absent in phagocyte NADPH-deficient murine infection.
(A) percentage of ΔagrBD (AIP-responsive in-frame deletion mutant carrying an intact RNAIII) cells and (B) bacterial burden in lung or spleen after 2 hr of intraperitoneal infection of wild-type (WT) C57BL/6 mice or phagocyte NADPH oxidase-deficient (Cybb-/-) mice (see Figure 9—figure supplement 1 for data with other organs). ΔagrBD and ΔrnaIII mutant cultures were grown separately and mixed at a 1:1 ratio either before (primed) or after (unprimed) overnight growth, as for Figure 3. Both primed and unprimed mixtures were diluted after overnight growth, grown to early log phase (OD600∼0.15), and used as inocula (1 × 108 CFU) for intraperitoneal infection (n=2 groups of 10 mice each). After 2 hr, lungs and spleen were harvested and homogenized; aliquots were diluted and plated to enumerate viable bacteria. Output ratios and total and mutant colony forming units (CFU) from tissue homogenates were determined as for Figure 4E and H. A Mann-Whitney test (panel 9 A) or Student’s two-tailed t-test (panel 9B) were used to determine the statistical significance of the difference between primed and unprimed cultures. Error bars indicate standard deviation (**p<0.01; ****p<0.0001).
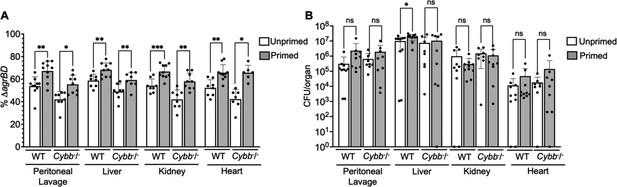
Long-lived protection by agr increases peritoneal fitness and dissemination to liver, kidney, and heart in both C57BL/6 mice and C57BL/6 Cybb-/- (gp91phox/nox2) mice.
(A) Percentage of ΔagrBD or (B) colony forming units (CFU) of S. aureus RN6734 ΔrnaIII (GAW183) and ΔagrBD mutant (GAW130) cells in the indicated organ 1 hr post intraperitoneal infection of wild-type (WT) C57BL/6 mice or phagocyte NADPH oxidase deficient (Cybb-/-) mice (see Figure 9 for data with lung and spleen). Wild-type and mutant strains were grown separately and mixed in a 1:1 ratio either before or after overnight growth, as for Figure 4. We called wild-type and mutant populations that were mixed prior to or after overnight growth; they were termed ‘primed’ and ‘unprimed,’ respectively. Both primed and unprimed mixtures were subsequently diluted, grown to early log phase (OD600∼0.15), and used as inoculum for intraperitoneal infection with 1 × 108 CFU (n=2 groups of 10 mice each). After 1 hr, the peritoneum was lavaged and the heart, kidneys, liver, lungs, and spleen (Figure 9) were harvested and homogenized. Samples were then diluted and plated to enumerate viable bacteria. Output ratios and total and mutant CFU from tissue homogenates were determined as for Figure 4E and H. A Mann-Whitney test (9 A) or Student’s two-tailed t-test (9B) were used to determine the statistical significance of the difference between primed and unprimed cultures. Error bars indicate standard deviation (**p<0.05; ****p<0.0001). Long-lived agr-mediated functions increased S. aureus pathogenesis in both wild-type and mutant mice, indicating a role for long-lived agr-mediated functions in pathogenesis other than protection from reactive oxygen species (ROS). Additionally, long-lived agr-mediated protection against ROS enhances fitness in lung and spleen (Figure 9), but it is dispensable for full virulence in other organs; protection is tissue-specific.

Schematic representation of agr-mediated protection from reactive oxygen species (ROS).
At low levels of oxidative stress, the redox sensor in AgrA binds to DNA at promoters P2 and P3, activating expression of the two operons. Expression of RNAIII blocks translation of Rot, which decreases respiration and production of superoxide. ROS quenchers (sodA and katA/ahpC) suppress formation of most ROS that would otherwise signal the redox sensor in AgrA to halt stimulation of RNAIII expression and the production of further superoxide via respiration. This feedback system regulates respiration thereby limiting the accumulation of ROS in wild-type cells. Wild-type cells are primed for induction of protective genes (e.g. clpB/C, dps) by loss of the rot repressor system via an unknown mechanism when cells experience damage from high levels of oxidative stress (experimentally introduced as lethal exogenous H2O2); Δagr cells that experience high levels of endogenous H2O2 fail to induce protective genes. Exogenous H2O2 or high levels of endogenous ROS, for example from extreme stress due to ciprofloxacin (Kumar et al., 2017), lower RNAIII expression and allow Rot to stimulate bsaA expression, which produces a protective antioxidant. The protective action of an ahpC deficiency acts through compensatory expression of katA, which results in more effective scavenging of H2O2 produced from increased respiration in Δagr strains and/or exogenous lethal H2O2.
© 2024, BioRender Inc. Figure 10 was created with BioRender.com and is published under a CC BY-NC-ND 4.0. Further reproductions must adhere to the terms of this license

Relationship of agr priming and virulence.
The ecology of abscess formation and subsequent bacterial dissemination can be described as a cycle. (a) During abscess formation, a hallmark of S. aureus disease, agr is activated by high bacterial cell density (quorum sensing) (Wright et al., 2005). (b) The bacterium assumes a primed stage due to repression of the rot repressor. (c, d, e) The lethal effects of immune challenge, which is called triggering (Andrade-Linares et al., 2016), are survived by the persistence (‘memory’) of the agr-activated state. (f) agr expression is inactivated by oxidation, thereby elevating expression of the antioxidant bsaA (Sun et al., 2012), which enables proliferation when oxidative stress is sublethal (Sun et al., 2012). (g) By surviving damage caused by lethal exogenous oxidative stress, primed S. aureus escape from the localized abscess to produce new infectious lesions (bloodstream dissemination) or to infect new hosts, where the cycle would be repeated.
© 2024, BioRender Inc. Figure 11 was created with BioRender.com and is published under a CC BY-NC-ND 4.0. Further reproductions must adhere to the terms of this license
Tables
Bacterial strains*.
| Strain | Background | Relevant genotype | Reference or source |
|---|---|---|---|
| BS819 | LAC | agr group I wild-type (CC8), ErmS | Boles et al., 2010 |
| BS1348 | BS819 | agr::tetM | Kumar et al., 2017 |
| BS820 | BS819 | agr::ermC | Kumar et al., 2017 |
| BS821 | BS819 | rnaIII::cadA | Wilde et al., 2015 |
| BS12 | Newman | agr group I wild-type (CC8) | Duthie and Lorenz, 1952 |
| BS13 | BS12 | agr::tetM | Geisinger et al., 2012 |
| BS669 | BS12 | rnaIII::cadA | Kumar et al., 2017 |
| BS39 | BS39 clinical strain | agr (+) clinical isolate (CC45) | Benson et al., 2011 |
| BS40 | BS40 clinical strain | agr (-) clinical isolate (CC45) | Benson et al., 2011 |
| BS867 | JE2 | agr group I wild-type (CC8) | Fey et al., 2013 |
| BS1010 | BS867 | agr::cadA in S. aureus JE2 | This study |
| BS1280 | BS12 | saeQRS::spec | Benson et al., 2012 |
| BS1282 | BS12 | agr::tetM, saeQRS::spec | Benson et al., 2012 |
| BS653 | E. coli | Top10 with pJC1111 (ampR in E. coli; CdR in S. aureus) | Chen et al., 2014 |
| BS656 | RN4220 | RN4220 with pRN7023 [shuttle vector (ampR in E. coli; CmR in S. aureus) containing SaPI1 int] | Chen et al., 2014 |
| BS435 | RN6734 | agr group-I prototype strain, derivative of NCTC 8325 | Ji et al., 1997 |
| BS688 | BS435 | agr::cadA | This study |
| GAW130 | BS435 | agr::cadA, SaPI1 attC::pGAW98 (agr-I ΔagrBD) | This study |
| GAW183 | BS435 | rnaIII::cad | This study |
| BS450 | MW2 | agr group I wild-type (CC1) | Baba et al., 2002 |
| BS451 | MW2 | agr::tetM | This study |
| BS988 | 126 a | agr (+) clinical isolate (CC5) | Benson et al., 2011 |
| BS989 | 127b | agr (-) clinical isolate (CC5) | Benson et al., 2011 |
| BS842 | BS819 | BS820 with SaPI1-attC::agr-IpJC1111 (agr-I, 8325–4) | Kumar et al., 2017 |
| BS1301 | BS819 | rot::Tn917 | This study |
| BS1302 | BS819 | agr::tetM, rot::Tn917 | This study |
| BS1279 | BS12 | rot::Tn917 | Benson et al., 2012 |
| BS1281 | BS12 | agr::tetM, rot::Tn917 | Benson et al., 2012 |
| VJT14.28 | BS12 | pOS1-Plgt-sodARBS-rot | Benson et al., 2012 |
| BS1486 | BS819 | ahpC::bursa (NE911) | This study, Fey et al., 2013 |
| BS1487 | BS819 | agr::tetM, ahpC::bursa | This study, Fey et al., 2013 |
| BS1488 | BS819 | katA::bursa (NE1366) | This study, Fey et al., 2013 |
| BS1489 | BS819 | agr::tetM, katA::bursa | This study, Fey et al., 2013 |
| BS1399 | BS12 | sodA::tetM, sodM::ermC | Kehl-Fie et al., 2011 |
| BS1422 | BS819 | sodA::tetM | This study |
| BS1423 | BS819 | agr::ermC, sodA::tetM | This study |
| BS1435 | BS819 | sigB clean deletion | Lauderdale et al., 2009 |
| BS1436 | BS819 | agr::tetM, sigB clean deletion | Lauderdale et al., 2009 |
| BS1246 | BS12 | mgrA::cat | Luong et al., 2006 |
| BS999 | BS819 | BS819 with SaPI1 attC::pGYlux (vector containing promoterless lux) | Mesak et al., 2009 |
| BS1222 | BS819 | BS819 with SaPI1 attC::Pagrp3-lux | Figueroa et al., 2014 |
| BS1518 | BS12 | agr::tetM, mgrA::cat | This study |
| BS1527 | BS867 | bshC::bursa (NE230) | Fey et al., 2013 |
| BS1528 | BS867 | agr::tetM, bshC::bursa | This study |
| BS1522 | BS867 | gpxA2::bursa (NE563) | Fey et al., 2013 |
| BS1523 | BS867 | agr::tetM, gpxA2::bursa | This study |
| BS1490 | BS819 | bsaA::bursa (NE1730) | This study |
| BS1491 | BS819 | bsaA::bursa, agr::tetM | This study |
| BS1707 | BS819 | BS1422 with SaPI-attC::PsarA-sodRBS-sodA | This study |
| BS1708 | BS819 | BS1348 with SaPI-attC::PsarA-sodRBS-sodA | This study |
| BS1494 | BS867 | ahpF::bursa (NE1571) | Fey et al., 2013 |
| BS1504 | BS867 | agr::tetM, ahpF::bursa | This study |
| BS1495 | BS867 | SAUSA300_0377::bursa (NE725) | Fey et al., 2013 |
| BS1501 | BS867 | agr::tetM, SAUSA300_0377::bursa | This study |
| BS1496 | BS867 | SAUSA300_0378::bursa (NE537) | Fey et al., 2013 |
| BS1502 | BS867 | agr::tetM, SAUSA300_0378::bursa | This study |
| BS1744 | BS819 | acnA::bursa (NE861) | This study |
| BS1745 | BS1348 | agr::tetM, acnA::bursa | This study |
-
*
All bacterial strains are S. aureus, unless otherwise indicated. Abbreviations: CC, clonal complex; NEx, strain designation in the Nebraska Transposon Mutant Library (Fey et al., 2013).
Oligonucleotides.
| # | Name | Gene/Target | Sequence 5’→ 3’ | Source |
|---|---|---|---|---|
| 1 | pflBRT.1a | pflB | AAAAATGGAAGATGGAACAGACAC | Kinkel et al., 2013 |
| 2 | pflBRT.1b | TCGATAACTGCATTACTTGTTCC | ||
| 3 | pflART.1a | pflA | TGACAAACATATTAGATTGACAGGAAAGC | Chen et al., 2009 |
| 4 | pflART.1b | ATCATCAGAATAACCAGGCACAAGG | ||
| 5 | ldh2RT.1a | ldh2 | GGATCTGTAGGATCAAGCTATGCC | Richardson et al., 2008 |
| 6 | ldh2RT.2b | TGGTGAAGGACTGTGGACTGTACC | ||
| 7 | nrdGRT.1a | nrdG | CAGTGTTTATGTATCAGGATGTCC | Kinkel et al., 2013 |
| 8 | nrdGRT.1b | GTTCGCCACCTAATAGACTTAGCC | ||
| 9 | qoxB-RT.3A | qoxB | GTTGTACTTGGCATGTTCGCC | Dmitriev et al., 2021 |
| 10 | qoxB-RT.3B | GGCATTATGGTGCATCTTACC | ||
| 11 | cydA-RT.1A | cydA | CATTTCGATACATCTTCCCATGCC | Dmitriev et al., 2021 |
| 12 | cydA-RT.1B | ATCTGCTAAGAAACTCAATAGTCC | ||
| 13 | hmp-RT.1A | hmp | TGACTTTAGTGAATTTACACCAGG | Dmitriev et al., 2021 |
| 14 | hmp-RT.1B | CGTTTAACGCCAAAAGTTAAATGG | ||
| 15 | spaRT1 | spA | CAAACCTGGTCAAGAACTTGTTGTTG | Brignoli et al., 2019 |
| 16 | spaRT2 | GCTAATGATAATCCACCAAATACAGTTG | ||
| 17 | clfB RT1 | clfA | GGATAGGCAATCATCAAGCACAAG | Brignoli et al., 2019 |
| 18 | clfB RT2 | GCTATCTACATTCGCACTGTTTGTG | ||
| 19 | ahp RT For | ahpC | CGTAAAAACCCTGGCGAAGTAT | Mashruwala and Boyd, 2017 |
| 20 | ahp RT Rev | TGCAATGTTTTAGCGCCTTCT | ||
| 21 | kat RT For | katA | TGGTGTTTTTGGGCATCCA | Shee et al., 2022 |
| 22 | kat RT Rev | CCCTAGGCCCTGCTGTCATA | ||
| 23 | rpoB F | rpoB | GAACATGCAACGTCAAGCAG | Dyzenhaus et al., 2023 |
| 24 | rpoB R | AATAGCCGCACCAGAATCAC | ||
| 25 | MPsodA#1 | sodA | AGGCGCGCCTTTATTTTGTTGCATTATATAATTCGTCAACTTTTTCCCAG | This study |
| 26 | MPsodA#2 | GGATGATTATTTATGGCTTTTGAATTACCAAAATTACCATACGC | This study | |
| 27 | MPsodA#3 | PsarA | TTCAAAAGCCATAAATAATCATCCTCCTAAGGTACCCGG | This study |
| 28 | MPsodA#4 | GCGGCCGCTCTGATATTTTTGACTAAACCAAATGCTAACCCAG | This study | |
| 29 | MPsodA#5 | pJC1111 | AAAATATCAGAGCGGCCGCCAG | This study |
| 30 | MPsodA#6 | ACAAAATAAAGGCGCGCCTATTCTAAATG | This study | |
| 31 | pJC1111 FOR | pJC1111-PsarA-sodRBS-sodA | TGGCCTTTTGCTCACATGTTCTTTCCTGCGTTATCCCCTGATTC | This study |
| 32 | pJC1111 REV | TGATATCAAAATTATACATGTCAACG | This study | |
| 33 | GWO#27 | agrBD | CAATTTTACACCACTCTCCTCACTGTCATTATACGATTTAG | This study |
| 34 | GWO#28 | TAATTTAAATAGAGAGTGTGATAGTAGGTGGAATTATTAAATAG | This study | |
| 35 | JCO#339 | agr flanking regions | GGTACCTGAAGCGGGCGAGCGAG | This study |
| 36 | JCO#340 | GGATCCGATAATAAAGTCAGTTAACGACGTATTCAATTGTAAATCTTGTTGG | This study | |
| 37 | JCO#342 | CTCGAGAAGAAGGGATGAGTTAATCATCATTATGAGAC | This study | |
| 38 | JCO#343 | GCATGCGATCTATCAAGGATGTGATGTTATGAAAGTCCAAATTTATCAATTACCG | This study |
Additional files
-
Supplementary file 1
RNA-seq comparison of agr wild-type and Δagr mutant strains grown to late exponential phase.
- https://cdn.elifesciences.org/articles/89098/elife-89098-supp1-v1.xlsx
-
Supplementary file 2
Data used for metabolic flux prediction.
- https://cdn.elifesciences.org/articles/89098/elife-89098-supp2-v1.xlsx
-
Supplementary file 3
RNA-seq comparison of ΔagrΔrot and Δagr mutant strains grown to early exponential phase, with or without treatment with H2O2.
- https://cdn.elifesciences.org/articles/89098/elife-89098-supp3-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/89098/elife-89098-mdarchecklist1-v1.docx





