Genetic screen identified PRMT5 as a neuroprotection target against cerebral ischemia
Figures
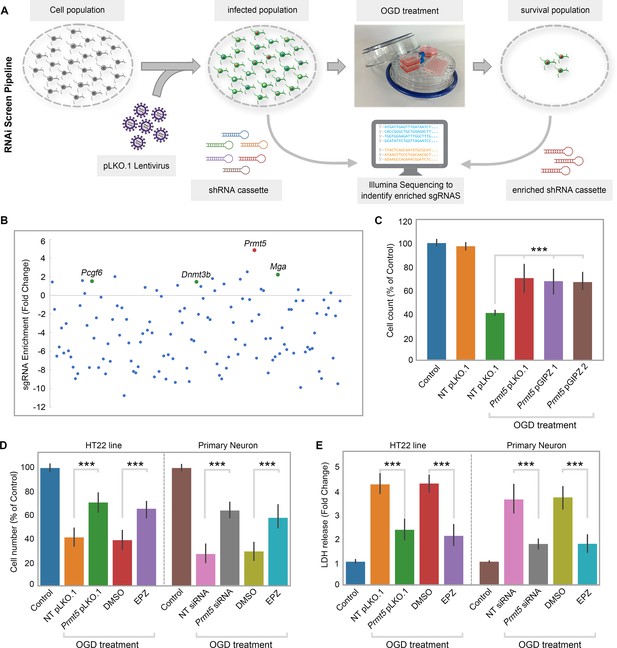
RNAi screen identified PRMT5 as a negative regulator of neuron survival after oxygen-glucose deprivation (OGD).
(A) Overview of the study methodology. (B) shRNA representation after OGD treatment. Prmt5 sgRNA have the highest enrichment after OGD treatment. (C) Effect of Prmt5 silencing in HT-22 cells after OGD. HT-22 cells (Control) or HT-22 cells transduced with the indicated shRNA lentiviruses were cultured with or without OGD treatment. Cell numbers were counted after the treatment and normalized against the control. Compared with non-targeting shRNA lentiviruses (NT pLKO.1), Prmt5 KD promoted a significant cell survival after OGD. (D–, E) Effect of Prmt5 silencing or PRMT5 inhibition in HT-22 cells and primary neurons after OGD. Cell viability was determined using the Cell Counting Kit-8. Cellular cytotoxicity was determined using the Lactate Dehydrogenase Activity Assay Kit. Compared with NT pLKO.1, Prmt5 pLKO.1 promoted a significant cell viability. Similar result could be found in PRMT5 inhibitor EPZ015666 (EPZ) vs. solvent control (DMSO). Graphing the mean with an SEM error bars, with ***p-value<0.001 by Student’s t-test (n = 5).
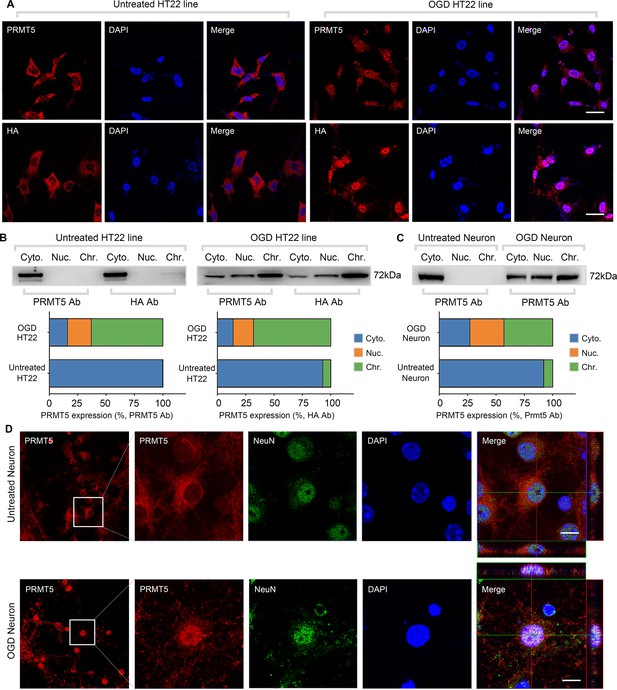
Oxygen-glucose deprivation led to PRMT5 nuclear translocation in vitro.
(A) Immunofluorescence staining to show PRMT5 cellular localization after oxygen-glucose deprivation (OGD) in HT-22 cells. HT-22 cells were stained with the PRMT5 antibody or HA antibody for C-terminal HA-tagging of the endogenous PRMT5 by CRIPR-mediated genome editing. Cell nuclei were counterstained by DAPI. Scale bar = 20 μm. (B) Biochemical fractionation to show PRMT5 cellular localization after OGD. PRMT5 in each fraction was detected by western blot using either the PRMT5 or HA antibody. (C, D) Primary cortical neurons were treated with or without OGD and subjected to biochemical fractionation. Primary neurons were stained with the PRMT5 or NeuN (neuronal marker) antibody. Scale bar = 8 μm. Cyto., Nuc., and Chr. stand for cytoplasmic, nucleoplasmic, and chromatin-bound fractions.
-
Figure 2—source data 1
Source data for the western blot analysis in the left panel of Figure 2B (anti-PRMT5 and anti-HA).
- https://cdn.elifesciences.org/articles/89754/elife-89754-fig2-data1-v1.zip
-
Figure 2—source data 2
Source data for the western blot analysis in the right panel of Figure 2B (anti-PRMT5 and anti-HA).
- https://cdn.elifesciences.org/articles/89754/elife-89754-fig2-data2-v1.zip
-
Figure 2—source data 3
Source data for the western blot analysis in Figure 2C (anti-PRMT5).
- https://cdn.elifesciences.org/articles/89754/elife-89754-fig2-data3-v1.zip
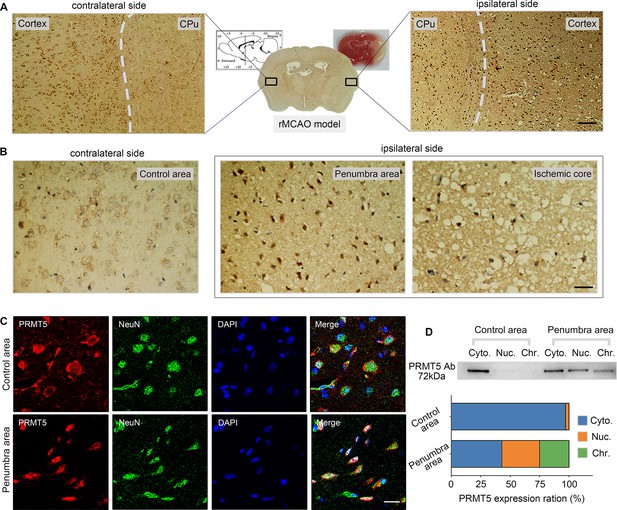
Middle cerebral artery occlusion led to PRMT5 nuclear translocation in vivo.
(A–C) Immunohistochemical and immunofluorescence staining to show PRMT5 cellular localization in ipsilateral and contralateral sides after middle cerebral artery occlusion (MCAO). Scale bar = 3 μm, 9 μm and 20 μm for A/B/C. (D) Biochemical fractionation to show PRMT5 cellular localization after MCAO. PRMT5 in each fraction was detected by western blot using the PRMT5 antibody. Cyto., Nuc., and Chr. stand for cytoplasmic, nucleoplasmic, and chromatin-bound fractions.
-
Figure 3—source data 1
Source data for the western blot analysis in Figure 3D (anti-PRMT5).
- https://cdn.elifesciences.org/articles/89754/elife-89754-fig3-data1-v1.zip
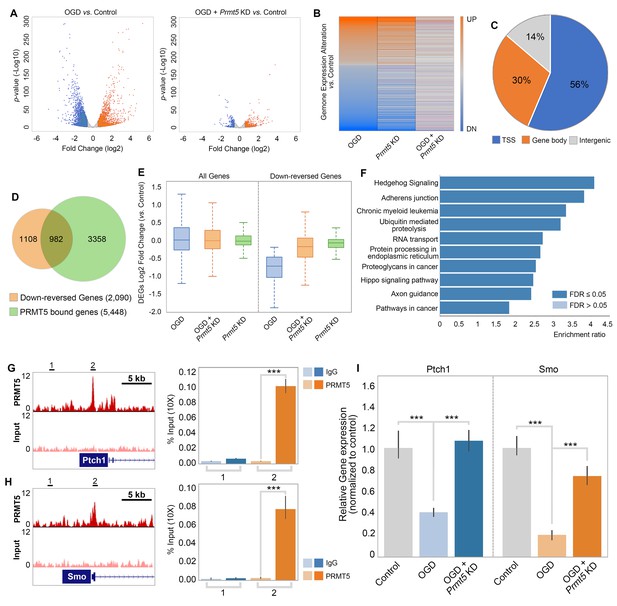
PRMT5 represses Hedgehog signaling expression after oxygen-glucose deprivation (OGD).
(A) Gene expression changes after OGD. HT-22 cells were transduced with Control (non-targeting) or Prmt5 shRNA lentiviruses, drug selected, and treated with OGD. Cells were collected and total RNAs were extracted for RNA-seq. Differentially expressed genes (DEGs) after OGD in the control cells were identified by log2-fold change >1 and p-value<0.01. Orange: upregulated genes after OGD; blue: downregulated genes after OGD. (B) Effect of Prmt5-KD on OGD-induced gene expression changes. OGD-induced DEGs were defined as described in (A), and the expression changes of the DEGs after OGD in control and Prmt5-KD cells were plotted by heat map. (C, D) Genomic regions occupied by PRMT5 based on ChIP-seq in HT-22 cells treated with OGD. Reversal of OGD-induced gene expression changes by Prmt5-KD. PRMT5-target genes were defined as described in the text. (E) Expression changes of all detected genes or PRMT5-target genes after OGD in control or Prmt5-KD cells were examined by box plots. (F) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of PRMT5-target genes. (G, H) Genome browser tracks of PRMT5 occupancy at selected PRMT5-target genes. ChIP-qPCR of PRMT5 occupancy in HT-22 cells treated with OGD at selected PRMT5-target genes (Ptch1 and Smo). (I) RT-qPCR to show the expression of selected PRMT5-target genes in control or Prmt5-KD HT-22 cells upon OGD treatment. Graphing the mean with an SEM error bars, with ***p-value<0.001 by Student’s t-test (n = 5).
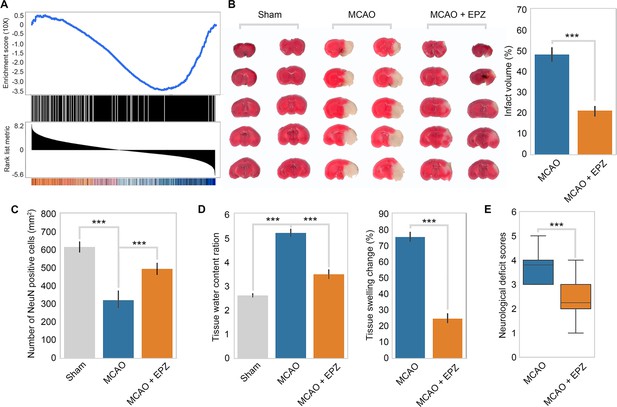
PRMT5 inhibition promotes neuron survival after ischemic injury in mouse brain.
(A) Gene set enrichment analysis for PRMT5-target genes in the middle cerebral artery occlusion (MCAO) mouse stroke model. Gene expression changes after MCAO was based on 29862458. (B) Effect of PRMT5 inhibition on infarct volume in the MCAO model. Mice were treated with subjected with sham (Sham group), MCAO with solvent (MCAO group), or MCAO with the PRMT5 inhibitor EPZ015666 (MCAO + EPZ group). The infarct volume was determined by 2,3,5-triphenyltetrazolium chloride (TTC) staining. (C–E) For tissue immunofluorescence staining, samples were fixed using 4% paraformaldehyde at room temperature for 15 min, followed by 0.5% Triton X-100 permeabilization for 10 min and 0.5% bovine serum albumin blocking for 30 min. The mice were sacrificed for NeuN immunofluorescence staining, and positive staining cell were count as survival neurons. Tissue water content ration, tissue swelling change, and neurological deficit score were determined as described in ‘Materials and methods’. Graphing the mean with an SEM error bars, with ***p-value<0.001 by Student’s t-test (n = 12).
Additional files
-
Supplementary file 1
A total of 125 genes involved in epigenetic and chromatin regulation were selected and used to generate an shRNA library in the pLKO.1 vector.
- https://cdn.elifesciences.org/articles/89754/elife-89754-supp1-v1.xlsx
-
Supplementary file 2
The PRMT5ChIP-seq were carried out in HT-22 cells treated with OGD, when intercepted with their RNA-seq data.
- https://cdn.elifesciences.org/articles/89754/elife-89754-supp2-v1.xlsx
-
Supplementary file 3
Primers and oligos used in this study.
- https://cdn.elifesciences.org/articles/89754/elife-89754-supp3-v1.xlsx
-
Supplementary file 4
Primary and secondary antibodies used in this study.
- https://cdn.elifesciences.org/articles/89754/elife-89754-supp4-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/89754/elife-89754-mdarchecklist1-v1.docx





