Gle1 is required for tRNA to stimulate Dbp5 ATPase activity in vitro and promote Dbp5-mediated tRNA export in vivo in Saccharomyces cerevisiae
Figures
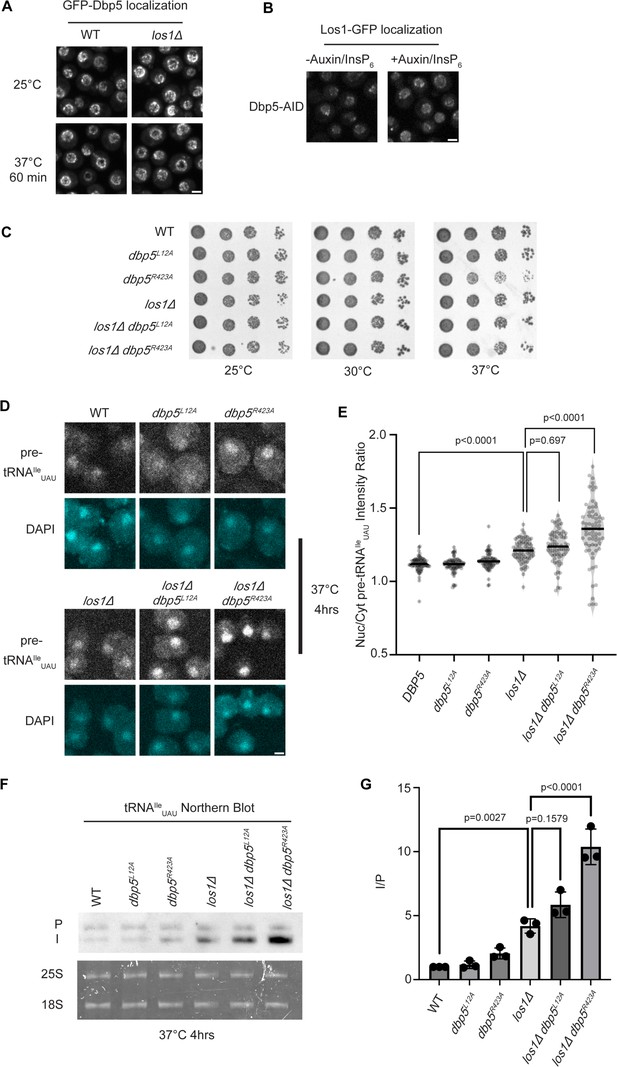
Dbp5 functions parallel to Los1 in pre-tRNA export.
(A) Fluorescent images show GFP-Dbp5 remains enriched at the nuclear periphery in wild-type and los1Δ at 25°C and 37°C. Scale bar represents 2 µm. (B) Los1-GFP remains nucleoplasmic and associated with nuclear periphery in Dbp5-AID after treatment with DMSO or 500 μM auxin and 10 μM InsP6 for 90 min. Scale bar represents 2 µm. (C) Spot assay for growth of strains containing untagged dbp5L12A, dbp5R423A, or los1Δ integrated at the endogenous gene locus after 2 d at 25, 30, and 37°C on YPD. (D) tRNA fluorescence in situ hybridization (FISH) targeting intron of tRNAIleUAU in indicated strains after pre-culture to early log phase at 25°C and shift to 37°C for 4 hr. Scale bar represents 2 µm. (E) Quantification of tRNA FISH from (E). Ratio of average nuclear to cytoplasmic pixel intensities was calculated across three independent replicate experiments and pooled for plotting. p-Values were calculated using one-way ANOVA. (F) Northern blot analysis targeting precursor and mature isoforms of tRNAIleUAU. Small RNAs were isolated from strains at mid-log phase growth after pre-culture at 25°C and shift to 37°C for 4 hr. ‘P’ bands represent intron-containing precursors that have 5′ leader/3′ trailer sequences, and ‘I’ bands represent intron-containing end-processed tRNA intermediates that have leader/trailer sequences removed. (G) Quantification of northern blot from (G). Ratio of signal from intron-containing end-processed intermediates (I) vs 5′ leader/3′ trailer-containing precursor (P) was calculated and presented relative to I/P ratio observed for WT. Error bars represent standard deviation, and p-values calculated using one-way ANOVA.
-
Figure 1—source data 1
Raw data files for northern blot in Figure 1F.
- https://cdn.elifesciences.org/articles/89835/elife-89835-fig1-data1-v1.zip
-
Figure 1—source data 2
Uncropped annotated raw northern blot for Figure 1F, with relevant bands highlighted.
- https://cdn.elifesciences.org/articles/89835/elife-89835-fig1-data2-v1.zip

Dbp5 functions parallel to Los1 in pre-tRNA export.
(A) dT fluorescence in situ hybridization (FISH) confirms induction of mRNA export defect and Dbp5 loss of function after addition of 500 μM auxin and 10 μM InsP6 in Dbp5-AID strain for 90 min. Scale bar represents 2 µm. (B) Spot assay for growth of shuffle strains containing untagged dbp5L12A or dbp5R423A on Leu-marked CEN plasmids in combination with los1Δ/msn5Δ. Genomic copy of Dbp5 has been replaced with His6Mx marker. Growth for 2 d at 25, 30, and 37°C on YPD was conducted following two rounds of counter selection for Ura marked WT Dbp5 CEN plasmids on 5′FOA. (C) tRNA FISH using a probe that can hybridize to the intron-containing and spliced isoform of tRNAIleUAU in indicated strains after pre-culture to early log phase at 25°C and shift to 37°C for 4 hr. Scale bar represents 2 µm. (D) tRNA FISH using a probe that can hybridize to the intron-containing and spliced isoform of tRNATyrGUA in indicated strains after pre-culture to early log phase at 25°C and shift to 37°C for 4 hr. Scale bar represents 2 µm. (E) Northern blot analysis targeting precursor and mature isoforms of tRNATyrGUA. Small RNAs were isolated from strains at mid-log phase growth after pre-culture at 25°C and shift to 37°C for 4 hr. ‘P’ bands represent intron-containing precursors that have 5′ leader/3′ trailer sequences, and ‘I’ bands represent intron-containing end-processed tRNA intermediates that have leader/trailer sequences removed. (F) Quantification of northern blot from (E). Ratio of signal from intron-containing end-processed intermediates (I) vs 5′ leader/3′ trailer-containing precursor (P) was calculated and presented relative to I/P ratio observed for WT. Error bars represent standard deviation, and p-values calculated using one-way ANOVA.
-
Figure 1—figure supplement 1—source data 1
Raw data files for northern blot in Figure 1—figure supplement 1E.
- https://cdn.elifesciences.org/articles/89835/elife-89835-fig1-figsupp1-data1-v1.zip
-
Figure 1—figure supplement 1—source data 2
Uncropped annotated raw northern blot for northern blot in Figure 1—figure supplement 1E.
- https://cdn.elifesciences.org/articles/89835/elife-89835-fig1-figsupp1-data2-v1.zip
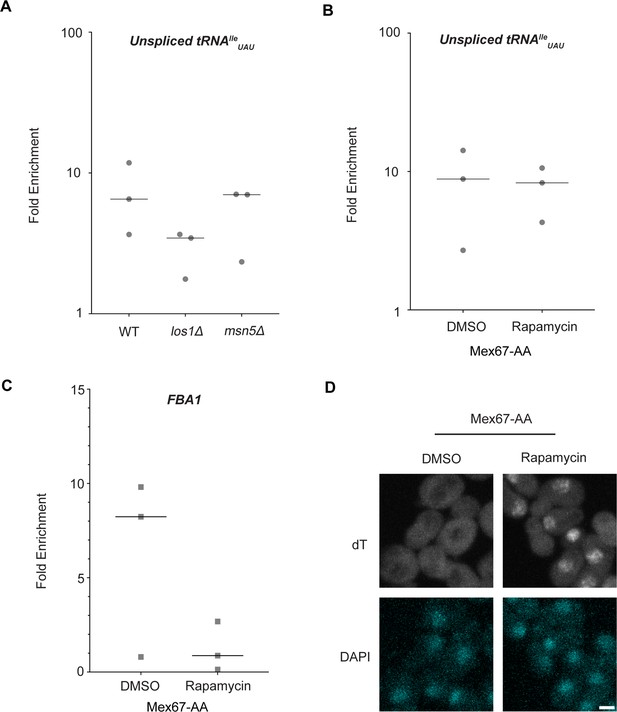
Los1 and Mex67 are not required for Dbp5 recruitment to pre-tRNAIleUAU.
(A) Plots show relative fold enrichment of tRNAIleUAU following prA-Dbp5 RNA IP in a wild-type (WT), los1Δ, and msn5Δ strain background. Abundance of target gene is normalized to abundance of the target transcript in input samples and represented as fold enrichment relative to RNA IP from an untagged control. (B) prA-Dbp5 RNA IP targeting tRNAIleUAU in Mex67-AA after either treatment with DMSO or 1 μg/ml rapamycin for 15 min. (C) prA-Dbp5 RNA IP targeting FBA1 mRNA in Mex67-AA after either treatment with DMSO or 1 μg/ml rapamycin for 15 min. (D) dT fluorescence in situ hybridization (FISH) confirming a mRNA export defect caused by loss of function of Mex67 following 15 min incubation with 1 μg/ml rapamycin compared to a DMSO control. Scale bar represents 2 µm.
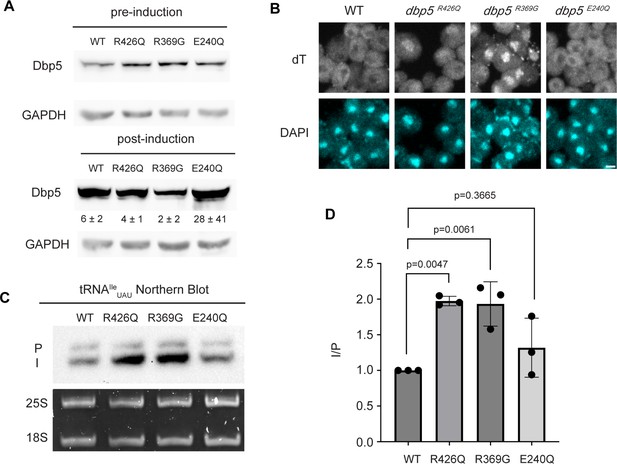
ATPase activity and Gle1 are required for Dbp5-mediated tRNA export in vivo.
(A) Western blot with either mouse monoclonal anti-DBP5 or anti-GAPDH (loading control) antibody to detect overexpression of untagged Dbp5 and Dbp5 ATPase mutants. Strains were cultured in raffinose at 25°C to derepress pGAL promoter (pre-induction), and expression was induced by shifting cultures into 2% galactose containing media for 6 hr (post-induction). Expression was stopped by addition of glucose for 1 hr. Overexpression was observed over the level of wild-type Dbp5 that is expressed from the endogenous locus and is present in the pre-induction samples. Top and bottom panels are from the same blot and processed equally. Quantification of the extent overexpression in post-induction samples is presented below Dbp5 bands. Signal intensity of Dbp5 bands was normalized to abundance of Gapdh and presented relative to pre-induction levels of expression. (B) dT fluorescence in situ hybridization (FISH) confirms previously reported mRNA export status phenotypes for Dbp5 ATPase mutants, with dbp5R426Q and dbp5R369G showing a nuclear accumulation of poly(A)-RNA. Scale bar represents 2 µm. (C) Northern blot analysis targeting precursor and mature isoforms of tRNAIleUAU from yeast strains overexpressing DBP5, dbp5R426Q, dbp5R369G, or dbp5E240Q. (D) Quantification of northern blot from (C). Ratio of signal from intron-containing end-processed intermediates (I) vs 5′ leader/3′ trailer-containing precursor (P) was calculated and presented relative to I/P ratio observed for WT. Error bars represent standard deviation, and p-values calculated using one-way ANOVA.
-
Figure 3—source data 1
Raw data files for western and northern blots in Figure 3A and C.
- https://cdn.elifesciences.org/articles/89835/elife-89835-fig3-data1-v1.zip
-
Figure 3—source data 2
Uncropped annotated raw western and northern blots for Figure 3A and C, with relevant bands highlighted.
- https://cdn.elifesciences.org/articles/89835/elife-89835-fig3-data2-v1.zip
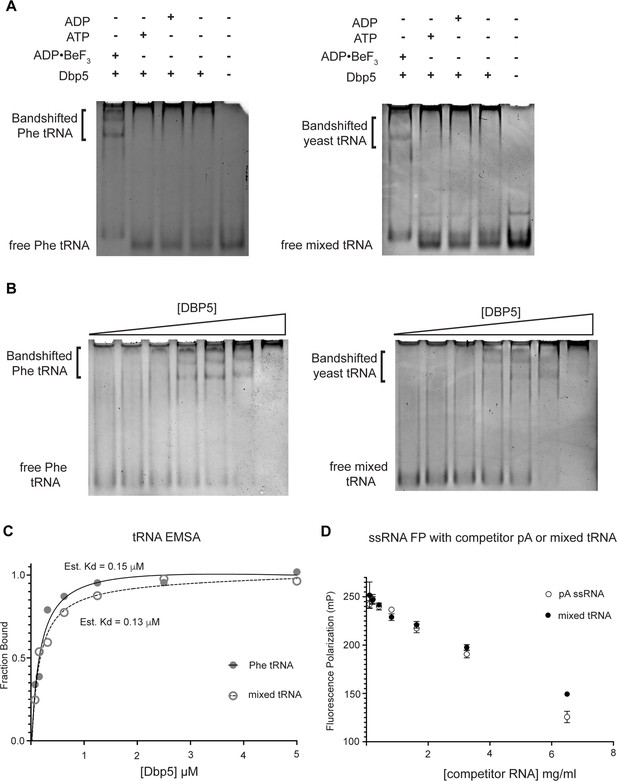
Dbp5 binds yeast tRNA in vitro.
(A) Full-length recombinant Dbp5 (fl-Dbp5) binds both mixed yeast tRNA and phenylalanine (Phe) tRNA in the presence of the ATP mimetic ADP•BeF3 by electrophoresis mobility shift assay (EMSA). RNA-binding reaction was conducted in the presence of ADP•BeF3, ATP, ADP, or no nucleotide and resolved on 6% native polyacrylamide gel at 70 V. Reactions contained 2 µM fl-Dbp5, 1 mM nucleotide when present, 250 ng tRNA, and binding buffer. (B) EMSA experiments in which increasing concentrations of fl-Dbp5 (1:2 dilution series starting at 5 uM Dbp5) were titrated into RNA-binding reactions in the presence of 1 mM ADP•BeF3, 250 ng mixed yeast tRNA or Phe tRNA, and binding buffer. (C) Band intensities of free probe and band shifts were quantified, and bound fraction was calculated for each well of EMSAs in (B). One site binding model was fit to the data using GraphPad Prism to estimate Kd for both mixed yeast tRNA and Phe tRNA. (D) Unlabeled pA ssRNA and mixed yeast tRNA compete with a 16nt fluorescein-labeled ssRNA for fl-Dbp5 binding. Fluorescence polarization competition assays were performed by titrating increasing concentration of an unlabeled competitor, pA (open square) or mixed yeast tRNA (closed circle), in reactions containing 50 nM fluorescein-labeled ssRNA, 2.5 mM ADP•BeF3, 1 µM Dbp5, and buffer. Error bars represent standard deviation of three independent experiments.
-
Figure 4—source data 1
Raw data files for electrophoretic mobility shift assays (EMSAs) in Figure 4A and B .
- https://cdn.elifesciences.org/articles/89835/elife-89835-fig4-data1-v1.zip
-
Figure 4—source data 2
Uncropped annotated electrophoretic mobility shift assays (EMSAs) for Figure 4A and B, with relevant bands highlighted.
- https://cdn.elifesciences.org/articles/89835/elife-89835-fig4-data2-v1.zip
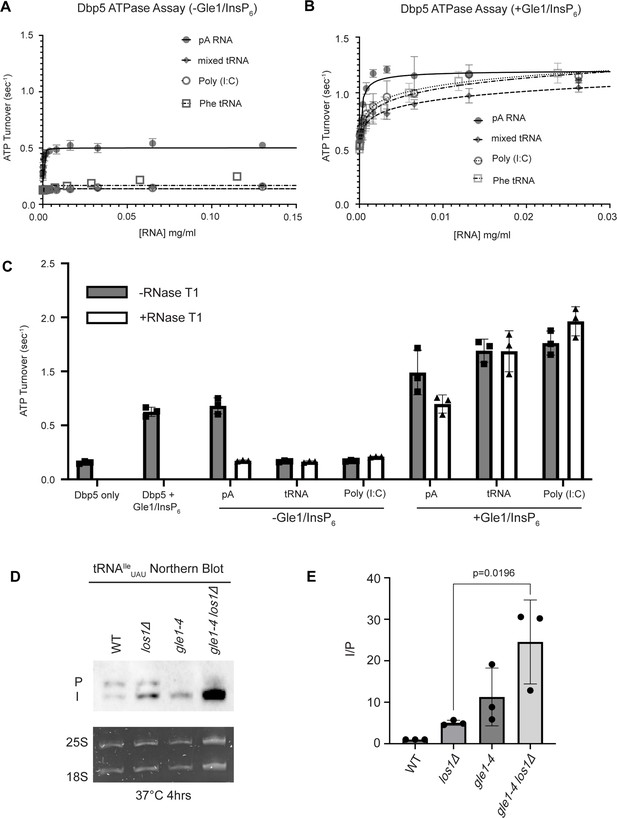
tRNA alone does not stimulate the Dbp5 ATPase cycle but can act synergistically with Gle1/InsP6 to fully activate Dbp5.
(A) ATPase activity of Dbp5 is stimulated by ssRNA (pA, closed square) but not mixed yeast tRNA (closed diamond), Phe tRNA (open square), or poly (I:C) dsRNA (open circle). Steady-state ATPase assays were conducted in the presence of 1 µM Dbp5, 2.5 mM ATP, and varying concentrations of RNA. Data was fit to Michaelis–Menten equation using GraphPad Prism. Error bars represent standard deviation of three independent experiments. (B) Gle1/InsP6 synergistically stimulates Dbp5 ATPase activity with mixed yeast tRNA (closed diamond), Phe tRNA (open square), and poly (I:C) dsRNA (open circle) substrates like ssRNA (pA). Steady-state ATPase assays were conducted in the presence of 1 µM Dbp5, 2.5 mM ATP, 2 µM Gle1, 2 µM InsP6, and varying concentration of RNA. Data was fit to an allosteric sigmoidal model in GraphPad Prism. Error bars represent standard deviation of three independent experiments. (C) RNase T1 treatment of RNA for 2 hr at 37°C prior to ATPase assays confirms that the observed synergistic activation of Dbp5 ATPase activity by Gle1/InsP6 and tRNA or dsRNA is not caused by low levels of contaminating ssRNA. Steady-state ATPase assays were conducted in the presence of 1 µM Dbp5, 2.5 mM ATP, 0.2 mg/ml RNA, and 2 µM Gle1/InsP6 when indicated. (D) Northern blot analysis targeting precursor and mature isoforms of tRNAIleUAU. Small RNAs were isolated from strains at mid-log phase growth after pre-culture at 25°C and shift to 37°C for 4 hr. ‘P’ bands represent intron-containing precursors that have 5′ leader/3′ trailer sequences, and ‘I’ bands represent intron-containing end-processed tRNA intermediates that have leader/trailer sequences removed. (E) Quantification of northern blot from (D). Ratio of signal from intron-containing end-processed intermediates (I) vs 5′ leader/3′ trailer-containing precursor (P) was calculated and presented relative to I/P ratio observed for WT. Error bars represent standard deviation, and p-values calculated using one-way ANOVA.
-
Figure 5—source data 1
Raw data files for northern blot in Figure 5D.
- https://cdn.elifesciences.org/articles/89835/elife-89835-fig5-data1-v1.zip
-
Figure 5—source data 2
Uncropped annotated raw northern blot from Figure 5D, with relevant bands highlighted.
- https://cdn.elifesciences.org/articles/89835/elife-89835-fig5-data2-v1.zip
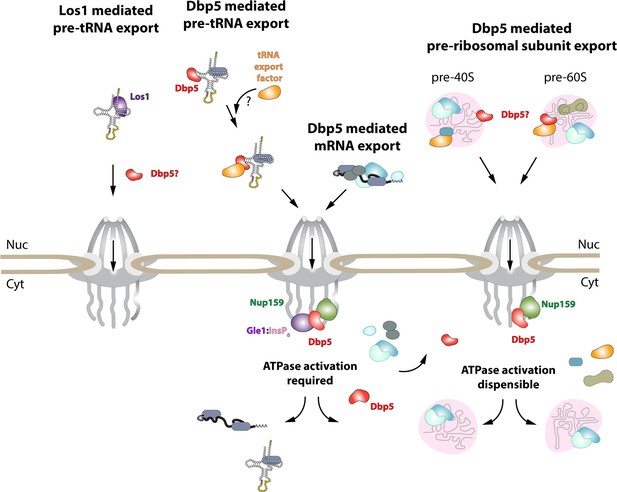
Model of Dbp5 function in mRNA, tRNA, and pre-ribosomal subunit export.
Data from this study support a model in which Dbp5-mediated export of both mRNA and tRNA require Gle1/InsP6 activation to remodel RNPs at the cytoplasmic face of the nuclear pore complex (NPC). For tRNA export, lack of RNA-mediated ATPase stimulation following RNA binding may lead to the formation of tRNA-bound intermediates that act as adapters for recruitment of yet to be identified transport factors. For mRNA export, it has been shown that Dbp5 function is limited to the nuclear periphery and Dbp5 does not form stable nuclear complexes with mRNA. Gle1/InsP6-mediated activation of Dbp5 catalytic cycle and RNA release likely promotes recycling of export factors and Dbp5 to function in further rounds of export or other functions. In contrast, for Dbp5-mediated pre-ribosomal subunit export Dbp5 ATPase cycle and Gle1/InsP6 stimulation are dispensable for transport. As such, RNA binding may promote export through an unknown mechanism and that Dbp5-mediated remodeling does not occur at NPCs, with factors such as Mex67 persisting on pre-ribosomal subunits following export to the cytoplasm. While a role for Dbp5 functioning in the Los1-mediated pre-tRNA export pathway cannot be excluded, the data presented in this study support a Dbp5-mediated tRNA export pathway that exists independent of and parallel to Los1.
Additional files
-
Supplementary file 1
Supplementary table of yeast strains.
List of yeast strains used in this study, along with associated genotypes and study of origin.
- https://cdn.elifesciences.org/articles/89835/elife-89835-supp1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/89835/elife-89835-mdarchecklist1-v1.pdf





